Lilian Krist
Can synthetic data reproduce real-world findings in epidemiology? A replication study using tree-based generative AI
Aug 19, 2025Abstract:Generative artificial intelligence for synthetic data generation holds substantial potential to address practical challenges in epidemiology. However, many current methods suffer from limited quality, high computational demands, and complexity for non-experts. Furthermore, common evaluation strategies for synthetic data often fail to directly reflect statistical utility. Against this background, a critical underexplored question is whether synthetic data can reliably reproduce key findings from epidemiological research. We propose the use of adversarial random forests (ARF) as an efficient and convenient method for synthesizing tabular epidemiological data. To evaluate its performance, we replicated statistical analyses from six epidemiological publications and compared original with synthetic results. These publications cover blood pressure, anthropometry, myocardial infarction, accelerometry, loneliness, and diabetes, based on data from the German National Cohort (NAKO Gesundheitsstudie), the Bremen STEMI Registry U45 Study, and the Guelph Family Health Study. Additionally, we assessed the impact of dimensionality and variable complexity on synthesis quality by limiting datasets to variables relevant for individual analyses, including necessary derivations. Across all replicated original studies, results from multiple synthetic data replications consistently aligned with original findings. Even for datasets with relatively low sample size-to-dimensionality ratios, the replication outcomes closely matched the original results across various descriptive and inferential analyses. Reducing dimensionality and pre-deriving variables further enhanced both quality and stability of the results.
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025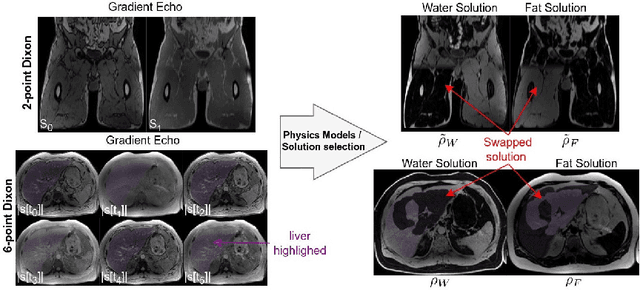
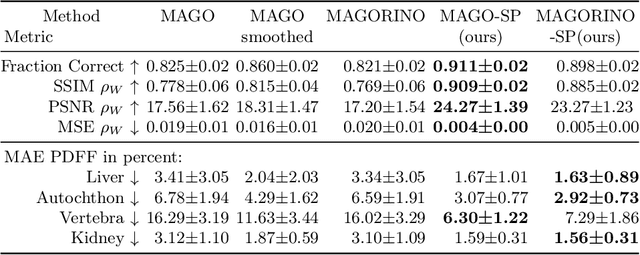
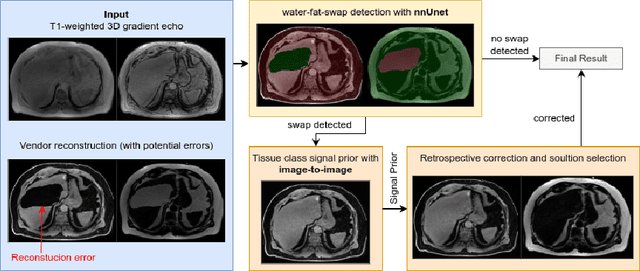
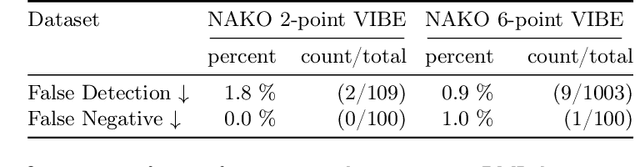
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge