Ishani Mondal
Intent Identification and Entity Extraction for Healthcare Queries in Indic Languages
Feb 19, 2023Abstract:Scarcity of data and technological limitations for resource-poor languages in developing countries like India poses a threat to the development of sophisticated NLU systems for healthcare. To assess the current status of various state-of-the-art language models in healthcare, this paper studies the problem by initially proposing two different Healthcare datasets, Indian Healthcare Query Intent-WebMD and 1mg (IHQID-WebMD and IHQID-1mg) and one real world Indian hospital query data in English and multiple Indic languages (Hindi, Bengali, Tamil, Telugu, Marathi and Gujarati) which are annotated with the query intents as well as entities. Our aim is to detect query intents and extract corresponding entities. We perform extensive experiments on a set of models in various realistic settings and explore two scenarios based on the access to English data only (less costly) and access to target language data (more expensive). We analyze context specific practical relevancy through empirical analysis. The results, expressed in terms of overall F1 score show that our approach is practically useful to identify intents and entities.
Explaining (Sarcastic) Utterances to Enhance Affect Understanding in Multimodal Dialogues
Nov 22, 2022Abstract:Conversations emerge as the primary media for exchanging ideas and conceptions. From the listener's perspective, identifying various affective qualities, such as sarcasm, humour, and emotions, is paramount for comprehending the true connotation of the emitted utterance. However, one of the major hurdles faced in learning these affect dimensions is the presence of figurative language, viz. irony, metaphor, or sarcasm. We hypothesize that any detection system constituting the exhaustive and explicit presentation of the emitted utterance would improve the overall comprehension of the dialogue. To this end, we explore the task of Sarcasm Explanation in Dialogues, which aims to unfold the hidden irony behind sarcastic utterances. We propose MOSES, a deep neural network, which takes a multimodal (sarcastic) dialogue instance as an input and generates a natural language sentence as its explanation. Subsequently, we leverage the generated explanation for various natural language understanding tasks in a conversational dialogue setup, such as sarcasm detection, humour identification, and emotion recognition. Our evaluation shows that MOSES outperforms the state-of-the-art system for SED by an average of ~2% on different evaluation metrics, such as ROUGE, BLEU, and METEOR. Further, we observe that leveraging the generated explanation advances three downstream tasks for affect classification - an average improvement of ~14% F1-score in the sarcasm detection task and ~2% in the humour identification and emotion recognition task. We also perform extensive analyses to assess the quality of the results.
Benchmarking Generalization via In-Context Instructions on 1,600+ Language Tasks
Apr 16, 2022
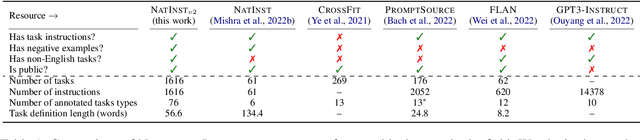
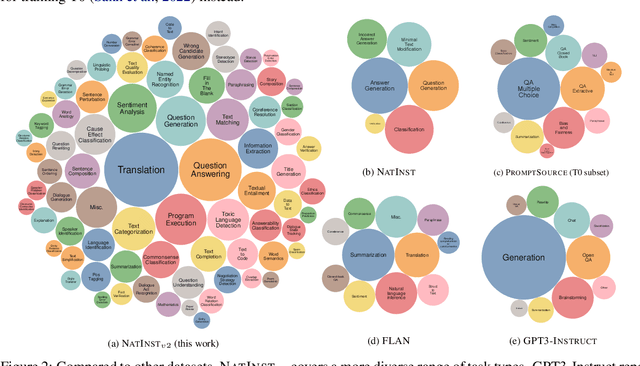
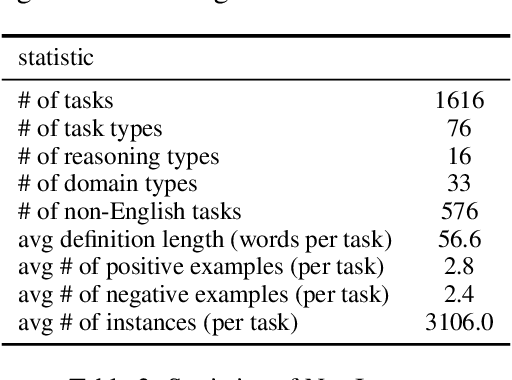
Abstract:How can we measure the generalization of models to a variety of unseen tasks when provided with their language instructions? To facilitate progress in this goal, we introduce Natural-Instructions v2, a collection of 1,600+ diverse language tasks and their expert written instructions. More importantly, the benchmark covers 70+ distinct task types, such as tagging, in-filling, and rewriting. This benchmark is collected with contributions of NLP practitioners in the community and through an iterative peer review process to ensure their quality. This benchmark enables large-scale evaluation of cross-task generalization of the models -- training on a subset of tasks and evaluating on the remaining unseen ones. For instance, we are able to rigorously quantify generalization as a function of various scaling parameters, such as the number of observed tasks, the number of instances, and model sizes. As a by-product of these experiments. we introduce Tk-Instruct, an encoder-decoder Transformer that is trained to follow a variety of in-context instructions (plain language task definitions or k-shot examples) which outperforms existing larger models on our benchmark. We hope this benchmark facilitates future progress toward more general-purpose language understanding models.
Global Readiness of Language Technology for Healthcare: What would it Take to Combat the Next Pandemic?
Apr 06, 2022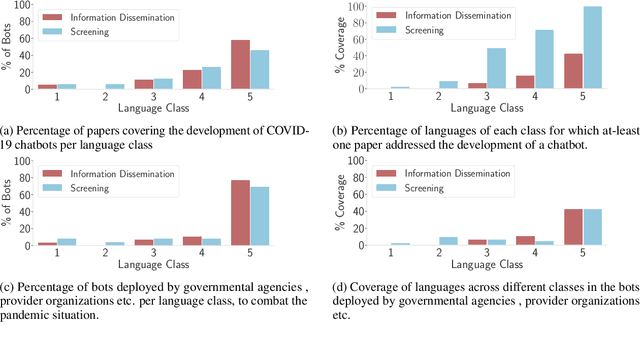
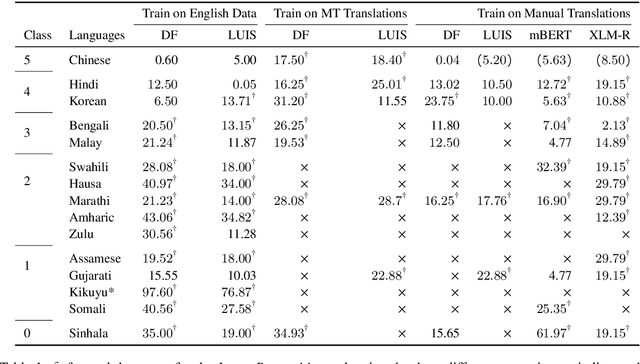
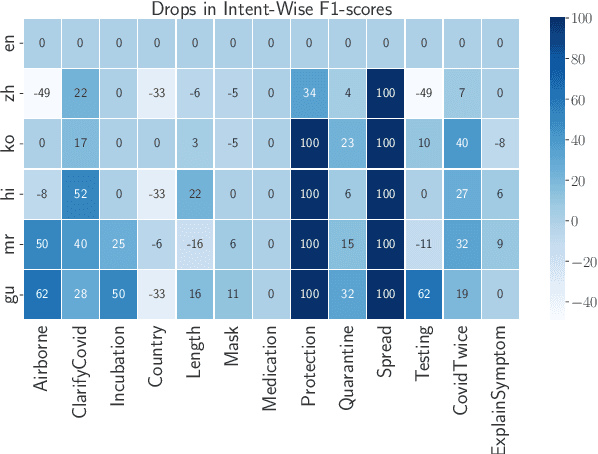

Abstract:The COVID-19 pandemic has brought out both the best and worst of language technology (LT). On one hand, conversational agents for information dissemination and basic diagnosis have seen widespread use, and arguably, had an important role in combating the pandemic. On the other hand, it has also become clear that such technologies are readily available for a handful of languages, and the vast majority of the global south is completely bereft of these benefits. What is the state of LT, especially conversational agents, for healthcare across the world's languages? And, what would it take to ensure global readiness of LT before the next pandemic? In this paper, we try to answer these questions through survey of existing literature and resources, as well as through a rapid chatbot building exercise for 15 Asian and African languages with varying amount of resource-availability. The study confirms the pitiful state of LT even for languages with large speaker bases, such as Sinhala and Hausa, and identifies the gaps that could help us prioritize research and investment strategies in LT for healthcare.
Multi-Objective Few-shot Learning for Fair Classification
Oct 05, 2021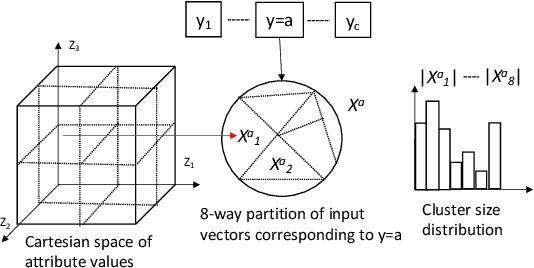
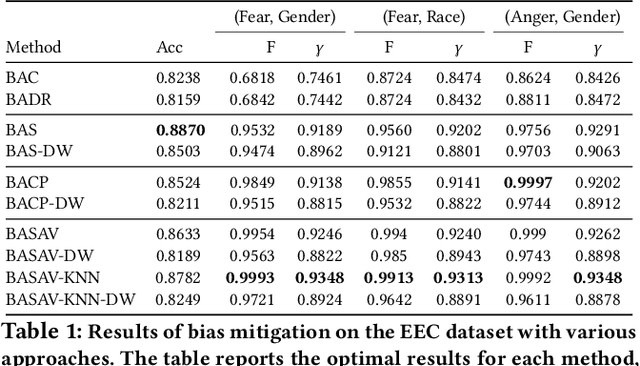
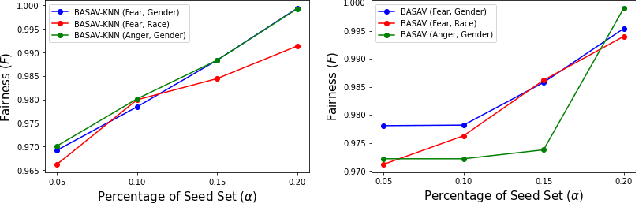
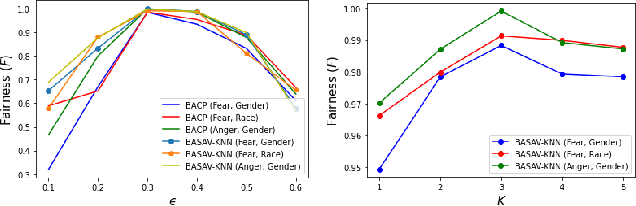
Abstract:In this paper, we propose a general framework for mitigating the disparities of the predicted classes with respect to secondary attributes within the data (e.g., race, gender etc.). Our proposed method involves learning a multi-objective function that in addition to learning the primary objective of predicting the primary class labels from the data, also employs a clustering-based heuristic to minimize the disparities of the class label distribution with respect to the cluster memberships, with the assumption that each cluster should ideally map to a distinct combination of attribute values. Experiments demonstrate effective mitigation of cognitive biases on a benchmark dataset without the use of annotations of secondary attribute values (the zero-shot case) or with the use of a small number of attribute value annotations (the few-shot case).
End-to-End NLP Knowledge Graph Construction
Jun 02, 2021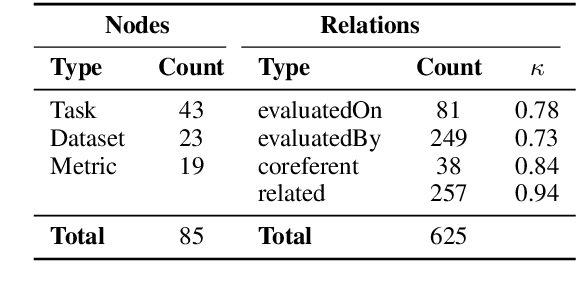
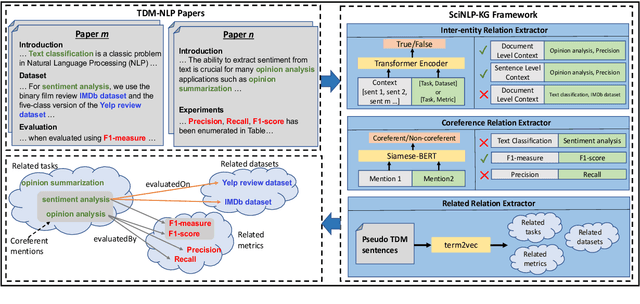
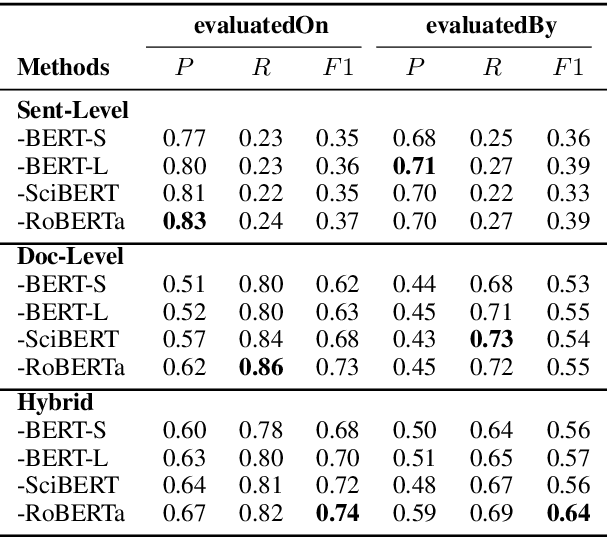
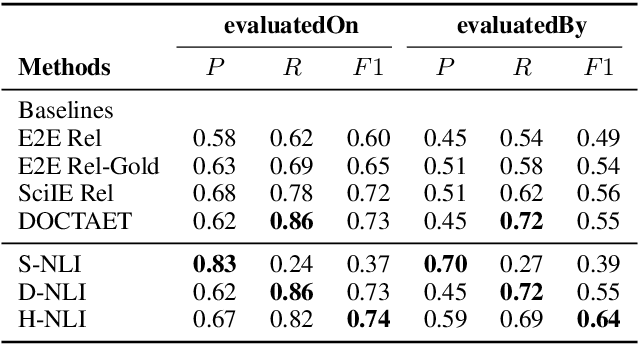
Abstract:This paper studies the end-to-end construction of an NLP Knowledge Graph (KG) from scientific papers. We focus on extracting four types of relations: evaluatedOn between tasks and datasets, evaluatedBy between tasks and evaluation metrics, as well as coreferent and related relations between the same type of entities. For instance, F1-score is coreferent with F-measure. We introduce novel methods for each of these relation types and apply our final framework (SciNLP-KG) to 30,000 NLP papers from ACL Anthology to build a large-scale KG, which can facilitate automatically constructing scientific leaderboards for the NLP community. The results of our experiments indicate that the resulting KG contains high-quality information.
BBAEG: Towards BERT-based Biomedical Adversarial Example Generation for Text Classification
Apr 05, 2021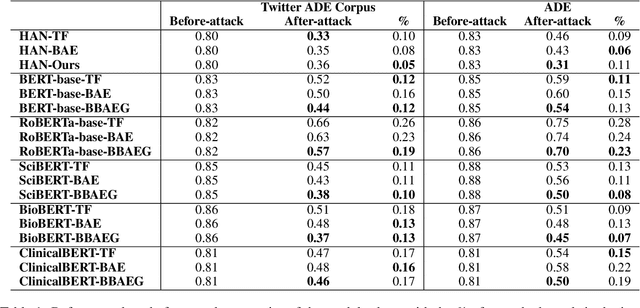

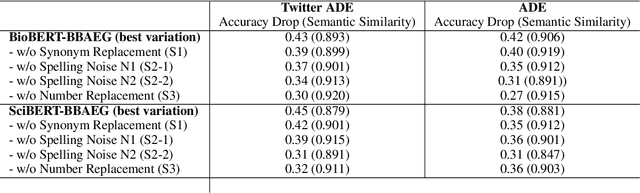
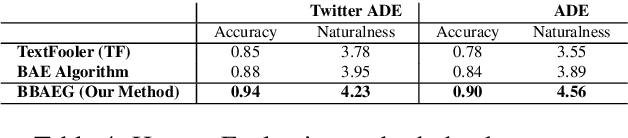
Abstract:Healthcare predictive analytics aids medical decision-making, diagnosis prediction and drug review analysis. Therefore, prediction accuracy is an important criteria which also necessitates robust predictive language models. However, the models using deep learning have been proven vulnerable towards insignificantly perturbed input instances which are less likely to be misclassified by humans. Recent efforts of generating adversaries using rule-based synonyms and BERT-MLMs have been witnessed in general domain, but the ever increasing biomedical literature poses unique challenges. We propose BBAEG (Biomedical BERT-based Adversarial Example Generation), a black-box attack algorithm for biomedical text classification, leveraging the strengths of both domain-specific synonym replacement for biomedical named entities and BERTMLM predictions, spelling variation and number replacement. Through automatic and human evaluation on two datasets, we demonstrate that BBAEG performs stronger attack with better language fluency, semantic coherence as compared to prior work.
Medical Entity Linking using Triplet Network
Dec 21, 2020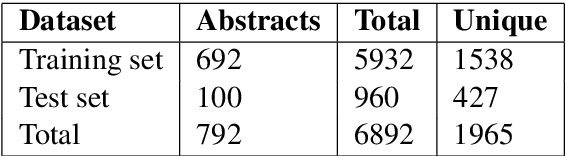
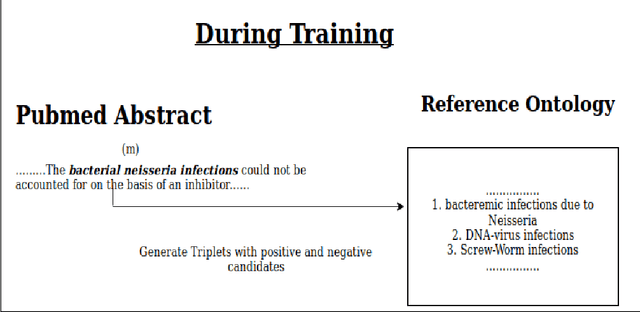
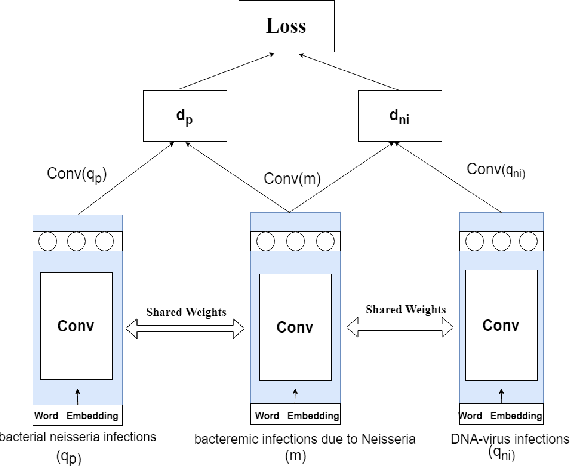
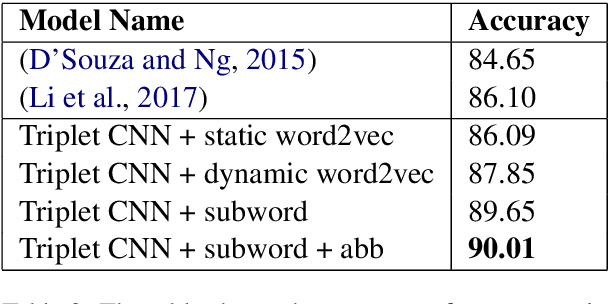
Abstract:Entity linking (or Normalization) is an essential task in text mining that maps the entity mentions in the medical text to standard entities in a given Knowledge Base (KB). This task is of great importance in the medical domain. It can also be used for merging different medical and clinical ontologies. In this paper, we center around the problem of disease linking or normalization. This task is executed in two phases: candidate generation and candidate scoring. In this paper, we present an approach to rank the candidate Knowledge Base entries based on their similarity with disease mention. We make use of the Triplet Network for candidate ranking. While the existing methods have used carefully generated sieves and external resources for candidate generation, we introduce a robust and portable candidate generation scheme that does not make use of the hand-crafted rules. Experimental results on the standard benchmark NCBI disease dataset demonstrate that our system outperforms the prior methods by a significant margin.
BERTChem-DDI : Improved Drug-Drug Interaction Prediction from text using Chemical Structure Information
Dec 21, 2020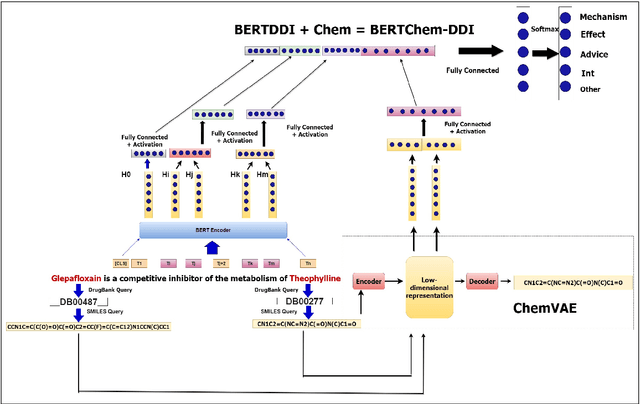
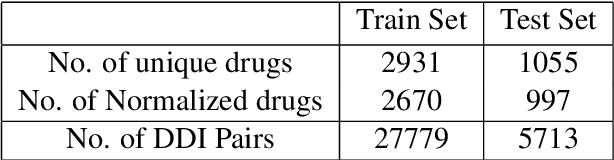
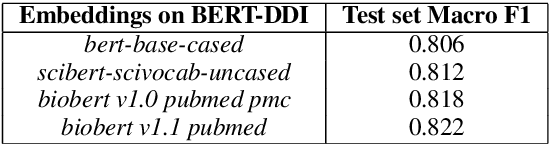
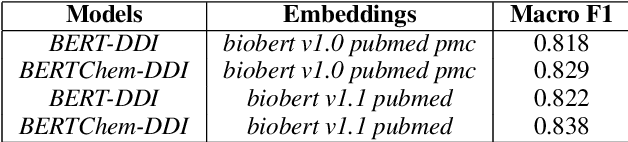
Abstract:Traditional biomedical version of embeddings obtained from pre-trained language models have recently shown state-of-the-art results for relation extraction (RE) tasks in the medical domain. In this paper, we explore how to incorporate domain knowledge, available in the form of molecular structure of drugs, for predicting Drug-Drug Interaction from textual corpus. We propose a method, BERTChem-DDI, to efficiently combine drug embeddings obtained from the rich chemical structure of drugs along with off-the-shelf domain-specific BioBERT embedding-based RE architecture. Experiments conducted on the DDIExtraction 2013 corpus clearly indicate that this strategy improves other strong baselines architectures by 3.4\% macro F1-score.
Towards Incorporating Entity-specific Knowledge Graph Information in Predicting Drug-Drug Interactions
Dec 21, 2020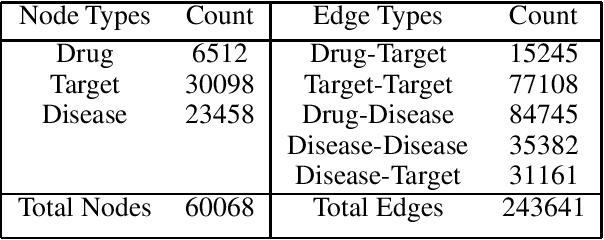
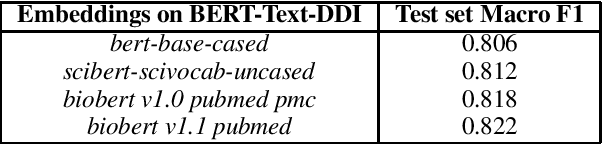
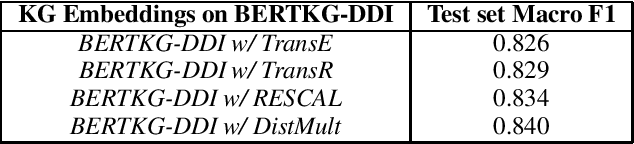

Abstract:Off-the-shelf biomedical embeddings obtained from the recently released various pre-trained language models (such as BERT, XLNET) have demonstrated state-of-the-art results (in terms of accuracy) for the various natural language understanding tasks (NLU) in the biomedical domain. Relation Classification (RC) falls into one of the most critical tasks. In this paper, we explore how to incorporate domain knowledge of the biomedical entities (such as drug, disease, genes), obtained from Knowledge Graph (KG) Embeddings, for predicting Drug-Drug Interaction from textual corpus. We propose a new method, BERTKG-DDI, to combine drug embeddings obtained from its interaction with other biomedical entities along with domain-specific BioBERT embedding-based RC architecture. Experiments conducted on the DDIExtraction 2013 corpus clearly indicate that this strategy improves other baselines architectures by 4.1% macro F1-score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge