Holger Roth
The Future of Digital Health with Federated Learning
Mar 18, 2020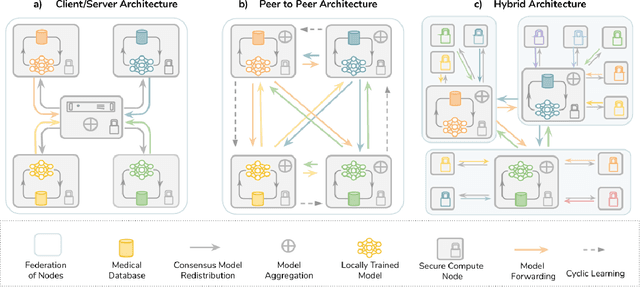
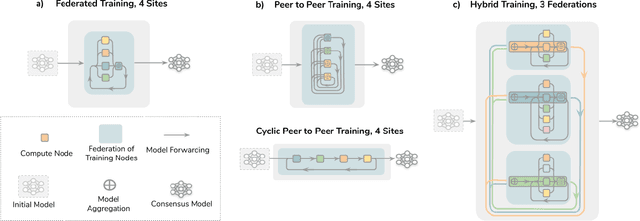
Abstract:Data-driven Machine Learning has emerged as a promising approach for building accurate and robust statistical models from medical data, which is collected in huge volumes by modern healthcare systems. Existing medical data is not fully exploited by ML primarily because it sits in data silos and privacy concerns restrict access to this data. However, without access to sufficient data, ML will be prevented from reaching its full potential and, ultimately, from making the transition from research to clinical practice. This paper considers key factors contributing to this issue, explores how Federated Learning (FL) may provide a solution for the future of digital health and highlights the challenges and considerations that need to be addressed.
C2FNAS: Coarse-to-Fine Neural Architecture Search for 3D Medical Image Segmentation
Dec 20, 2019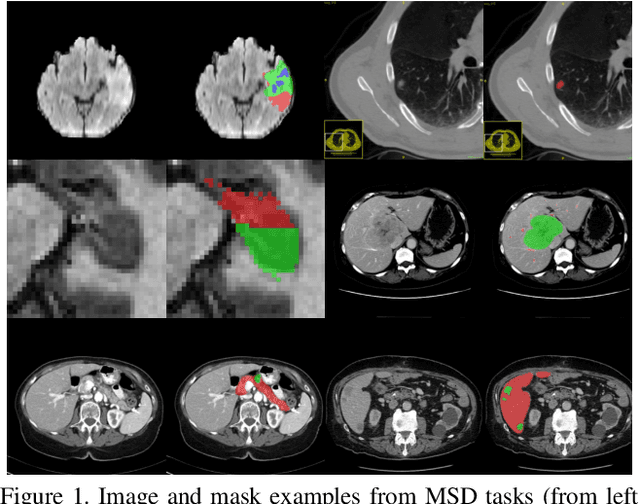
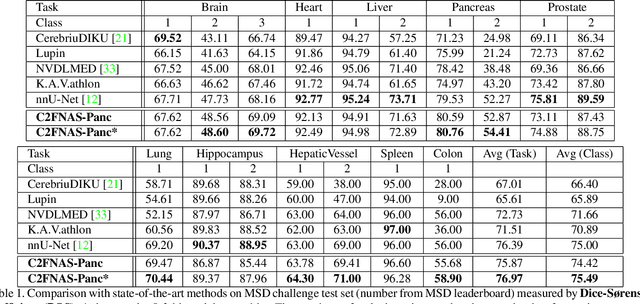

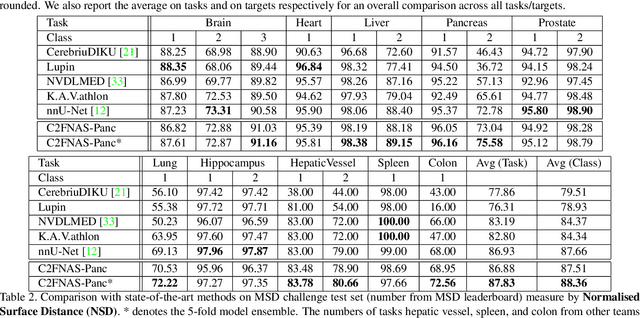
Abstract:3D convolution neural networks (CNN) have been proved very successful in parsing organs or tumours in 3D medical images, but it remains sophisticated and time-consuming to choose or design proper 3D networks given different task contexts. Recently, Neural Architecture Search (NAS) is proposed to solve this problem by searching for the best network architecture automatically. However, the inconsistency between search stage and deployment stage often exists in NAS algorithms due to memory constraints and large search space, which could become more serious when applying NAS to some memory and time consuming tasks, such as 3D medical image segmentation. In this paper, we propose coarse-to-fine neural architecture search (C2FNAS) to automatically search a 3D segmentation network from scratch without inconsistency on network size or input size. Specifically, we divide the search procedure into two stages: 1) the coarse stage, where we search the macro-level topology of the network, i.e. how each convolution module is connected to other modules; 2) the fine stage, where we search at micro-level for operations in each cell based on previous searched macro-level topology. The coarse-to-fine manner divides the search procedure into two consecutive stages and meanwhile resolves the inconsistency. We evaluate our method on 10 public datasets from Medical Segmentation Decalthon (MSD) challenge, and achieve state-of-the-art performance with the network searched using one dataset, which demonstrates the effectiveness and generalization of our searched models.
End-to-End Adversarial Shape Learning for Abdomen Organ Deep Segmentation
Oct 15, 2019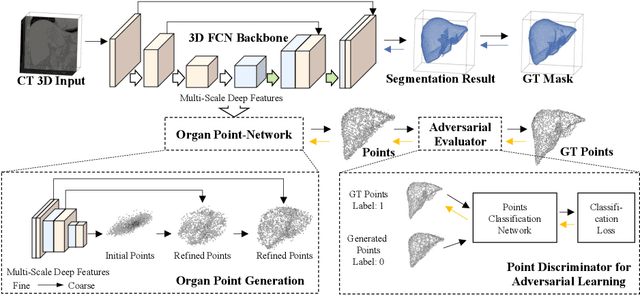
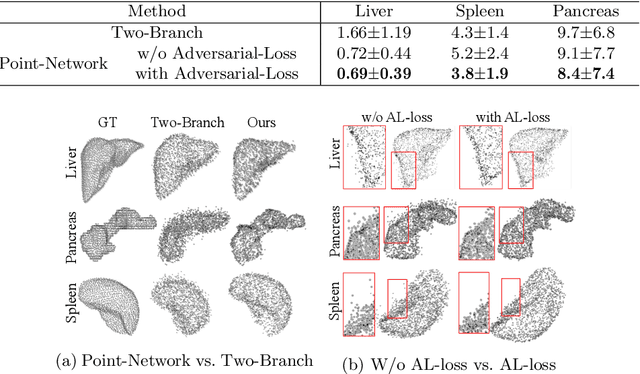
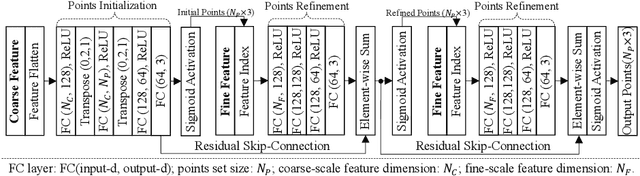
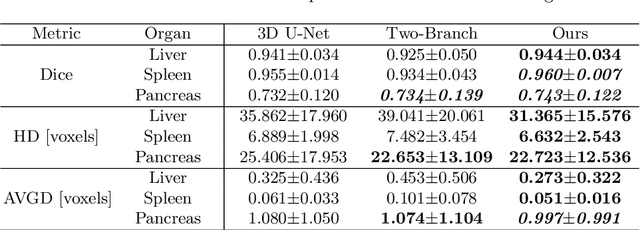
Abstract:Automatic segmentation of abdomen organs using medical imaging has many potential applications in clinical workflows. Recently, the state-of-the-art performance for organ segmentation has been achieved by deep learning models, i.e., convolutional neural network (CNN). However, it is challenging to train the conventional CNN-based segmentation models that aware of the shape and topology of organs. In this work, we tackle this problem by introducing a novel end-to-end shape learning architecture -- organ point-network. It takes deep learning features as inputs and generates organ shape representations as points that located on organ surface. We later present a novel adversarial shape learning objective function to optimize the point-network to capture shape information better. We train the point-network together with a CNN-based segmentation model in a multi-task fashion so that the shared network parameters can benefit from both shape learning and segmentation tasks. We demonstrate our method with three challenging abdomen organs including liver, spleen, and pancreas. The point-network generates surface points with fine-grained details and it is found critical for improving organ segmentation. Consequently, the deep segmentation model is improved by the introduced shape learning as significantly better Dice scores are observed for spleen and pancreas segmentation.
NeurReg: Neural Registration and Its Application to Image Segmentation
Oct 04, 2019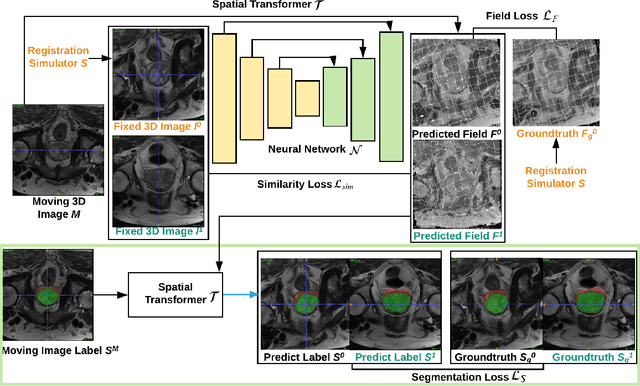
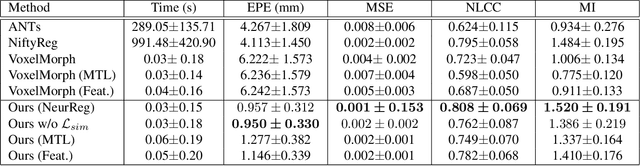
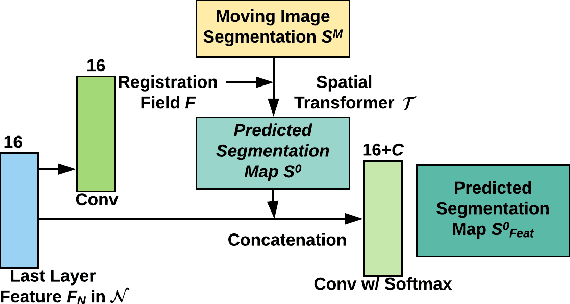
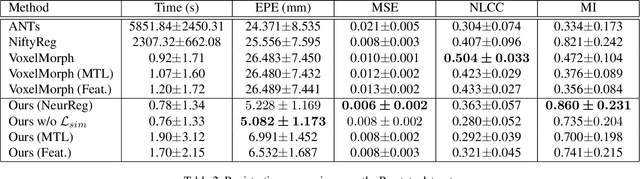
Abstract:Registration is a fundamental task in medical image analysis which can be applied to several tasks including image segmentation, intra-operative tracking, multi-modal image alignment, and motion analysis. Popular registration tools such as ANTs and NiftyReg optimize an objective function for each pair of images from scratch which is time-consuming for large images with complicated deformation. Facilitated by the rapid progress of deep learning, learning-based approaches such as VoxelMorph have been emerging for image registration. These approaches can achieve competitive performance in a fraction of a second on advanced GPUs. In this work, we construct a neural registration framework, called NeurReg, with a hybrid loss of displacement fields and data similarity, which substantially improves the current state-of-the-art of registrations. Within the framework, we simulate various transformations by a registration simulator which generates fixed image and displacement field ground truth for training. Furthermore, we design three segmentation frameworks based on the proposed registration framework: 1) atlas-based segmentation, 2) joint learning of both segmentation and registration tasks, and 3) multi-task learning with atlas-based segmentation as an intermediate feature. Extensive experimental results validate the effectiveness of the proposed NeurReg framework based on various metrics: the endpoint error (EPE) of the predicted displacement field, mean square error (MSE), normalized local cross-correlation (NLCC), mutual information (MI), Dice coefficient, uncertainty estimation, and the interpretability of the segmentation. The proposed NeurReg improves registration accuracy with fast inference speed, which can greatly accelerate related medical image analysis tasks.
Cardiac Segmentation of LGE MRI with Noisy Labels
Oct 02, 2019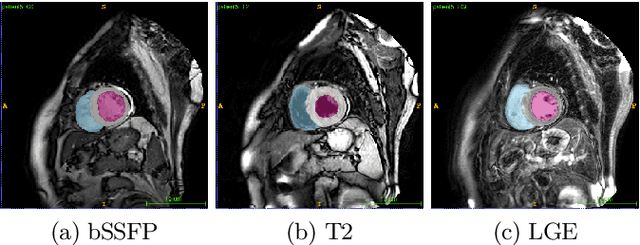
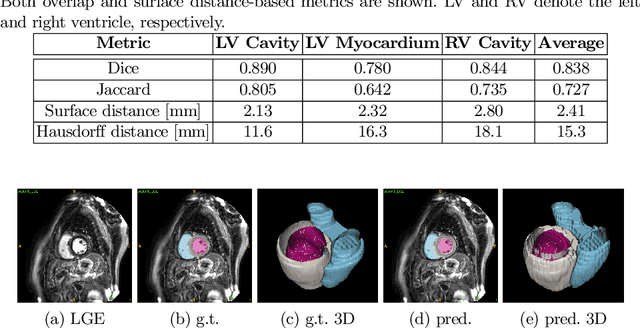
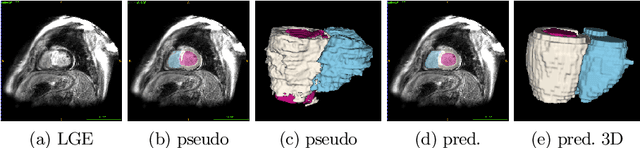
Abstract:In this work, we attempt the segmentation of cardiac structures in late gadolinium-enhanced (LGE) magnetic resonance images (MRI) using only minimal supervision in a two-step approach. In the first step, we register a small set of five LGE cardiac magnetic resonance (CMR) images with ground truth labels to a set of 40 target LGE CMR images without annotation. Each manually annotated ground truth provides labels of the myocardium and the left ventricle (LV) and right ventricle (RV) cavities, which are used as atlases. After multi-atlas label fusion by majority voting, we possess noisy labels for each of the targeted LGE images. A second set of manual labels exists for 30 patients of the target LGE CMR images, but are annotated on different MRI sequences (bSSFP and T2-weighted). Again, we use multi-atlas label fusion with a consistency constraint to further refine our noisy labels if additional annotations in other modalities are available for a given patient. In the second step, we train a deep convolutional network for semantic segmentation on the target data while using data augmentation techniques to avoid over-fitting to the noisy labels. After inference and simple post-processing, we achieve our final segmentation for the targeted LGE CMR images, resulting in an average Dice of 0.890, 0.780, and 0.844 for LV cavity, LV myocardium, and RV cavity, respectively.
Weakly supervised segmentation from extreme points
Oct 02, 2019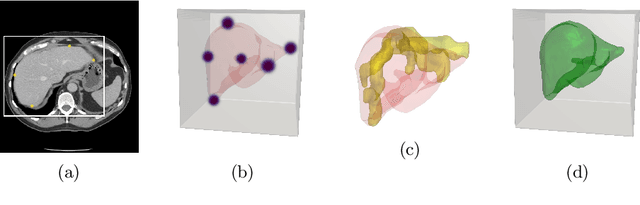
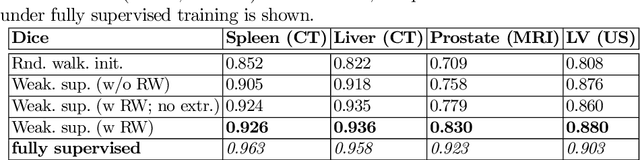
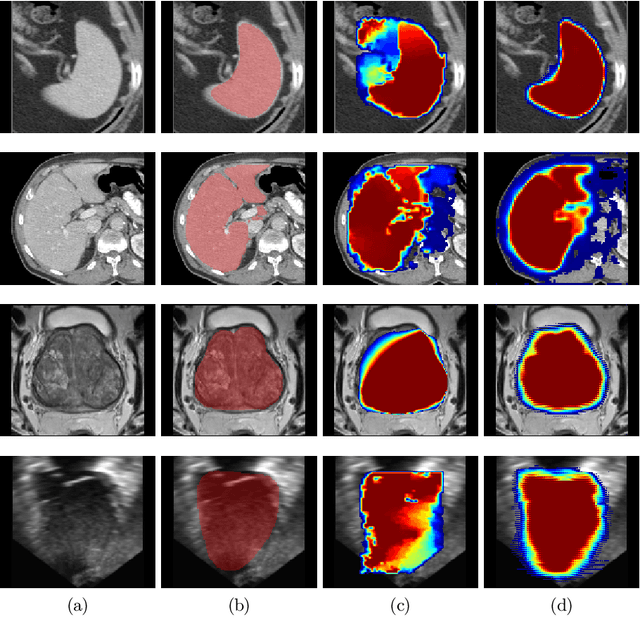
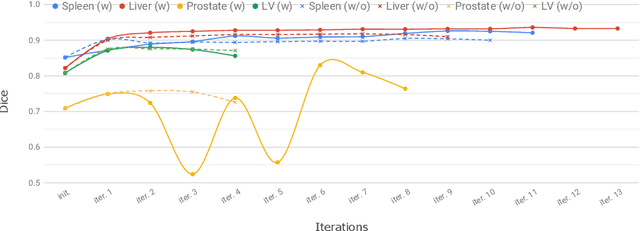
Abstract:Annotation of medical images has been a major bottleneck for the development of accurate and robust machine learning models. Annotation is costly and time-consuming and typically requires expert knowledge, especially in the medical domain. Here, we propose to use minimal user interaction in the form of extreme point clicks in order to train a segmentation model that can, in turn, be used to speed up the annotation of medical images. We use extreme points in each dimension of a 3D medical image to constrain an initial segmentation based on the random walker algorithm. This segmentation is then used as a weak supervisory signal to train a fully convolutional network that can segment the organ of interest based on the provided user clicks. We show that the network's predictions can be refined through several iterations of training and prediction using the same weakly annotated data. Ultimately, our method has the potential to speed up the generation process of new training datasets for the development of new machine learning and deep learning-based models for, but not exclusively, medical image analysis.
Correlation via synthesis: end-to-end nodule image generation and radiogenomic map learning based on generative adversarial network
Jul 08, 2019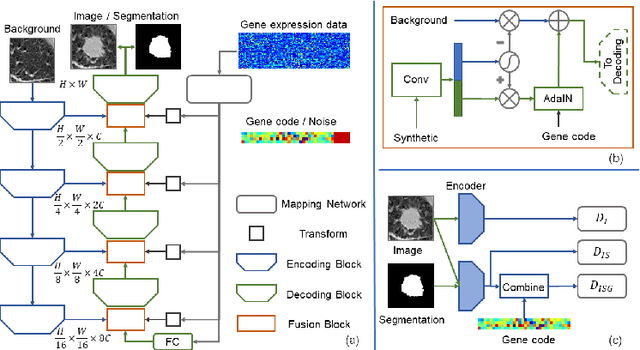

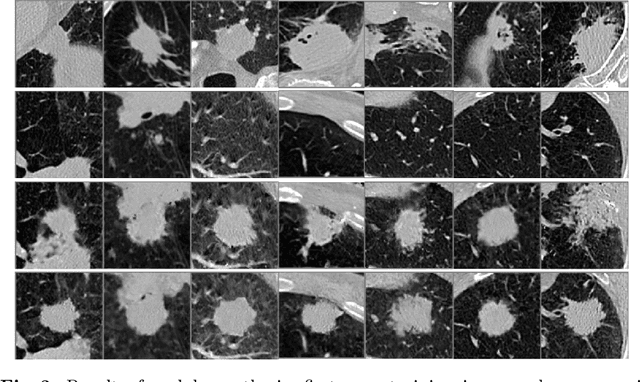
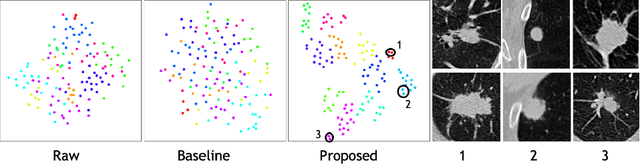
Abstract:Radiogenomic map linking image features and gene expression profiles is useful for noninvasively identifying molecular properties of a particular type of disease. Conventionally, such map is produced in three separate steps: 1) gene-clustering to "metagenes", 2) image feature extraction, and 3) statistical correlation between metagenes and image features. Each step is independently performed and relies on arbitrary measurements. In this work, we investigate the potential of an end-to-end method fusing gene data with image features to generate synthetic image and learn radiogenomic map simultaneously. To achieve this goal, we develop a generative adversarial network (GAN) conditioned on both background images and gene expression profiles, synthesizing the corresponding image. Image and gene features are fused at different scales to ensure the realism and quality of the synthesized image. We tested our method on non-small cell lung cancer (NSCLC) dataset. Results demonstrate that the proposed method produces realistic synthetic images, and provides a promising way to find gene-image relationship in a holistic end-to-end manner.
4D CNN for semantic segmentation of cardiac volumetric sequences
Jun 17, 2019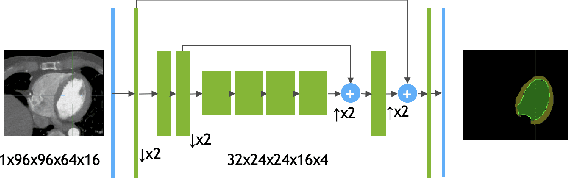
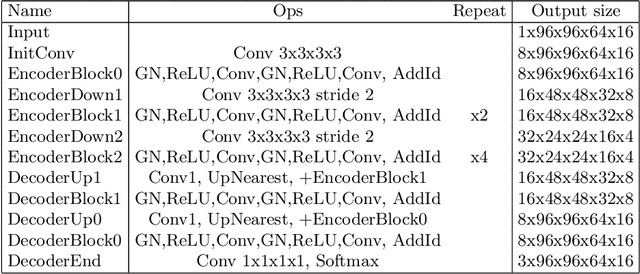
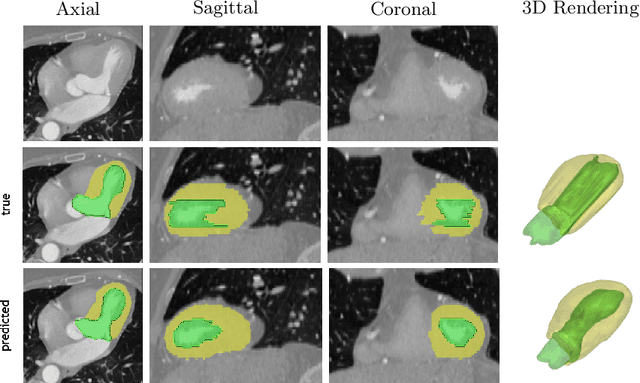
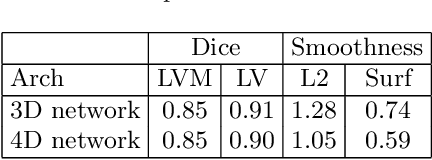
Abstract:We propose a 4D convolutional neural network (CNN) for the segmentation of retrospective ECG-gated cardiac CT, a series of single-channel volumetric data over time. While only a small subset of volumes in the temporal sequence are annotated, we define a sparse loss function on available labels to allow the network to leverage unlabeled images during training and generate a fully segmented sequence. We investigate the accuracy of the proposed 4D network to predict temporally consistent segmentations and compare with traditional 3D segmentation approaches. We demonstrate the feasibility of the 4D CNN and establish its performance on cardiac 4D CCTA.
When Unseen Domain Generalization is Unnecessary? Rethinking Data Augmentation
Jun 12, 2019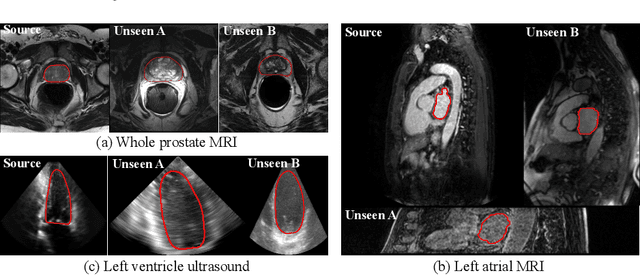
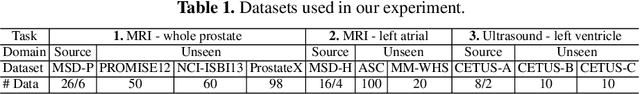
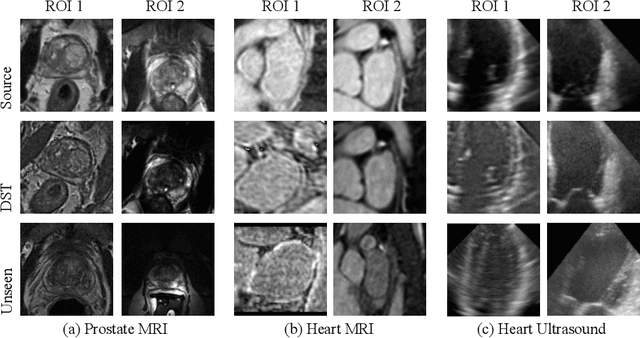
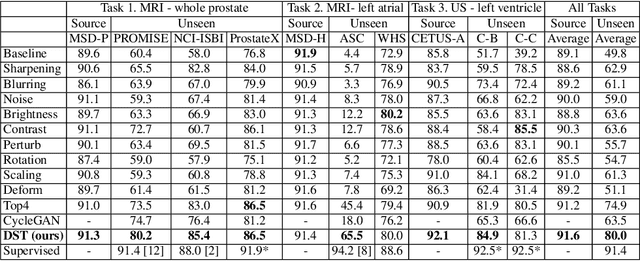
Abstract:Recent advances in deep learning for medical image segmentation demonstrate expert-level accuracy. However, in clinically realistic environments, such methods have marginal performance due to differences in image domains, including different imaging protocols, device vendors and patient populations. Here we consider the problem of domain generalization, when a model is trained once, and its performance generalizes to unseen domains. Intuitively, within a specific medical imaging modality the domain differences are smaller relative to natural images domain variability. We rethink data augmentation for medical 3D images and propose a deep stacked transformations (DST) approach for domain generalization. Specifically, a series of n stacked transformations are applied to each image in each mini-batch during network training to account for the contribution of domain-specific shifts in medical images. We comprehensively evaluate our method on three tasks: segmentation of whole prostate from 3D MRI, left atrial from 3D MRI, and left ventricle from 3D ultrasound. We demonstrate that when trained on a small source dataset, (i) on average, DST models on unseen datasets degrade only by 11% (Dice score change), compared to the conventional augmentation (degrading 39%) and CycleGAN-based domain adaptation method (degrading 25%); (ii) when evaluation on the same domain, DST is also better albeit only marginally. (iii) When training on large-sized data, DST on unseen domains reaches performance of state-of-the-art fully supervised models. These findings establish a strong benchmark for the study of domain generalization in medical imaging, and can be generalized to the design of robust deep segmentation models for clinical deployment.
Interactive segmentation of medical images through fully convolutional neural networks
Mar 19, 2019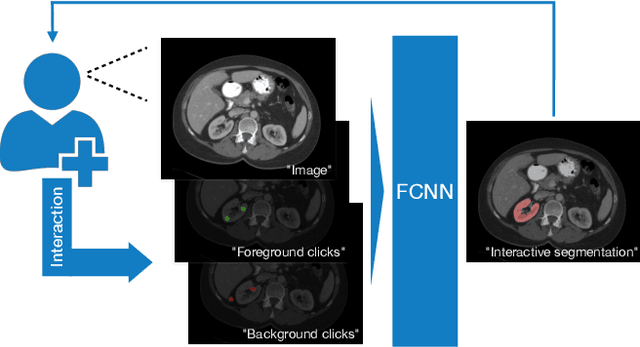
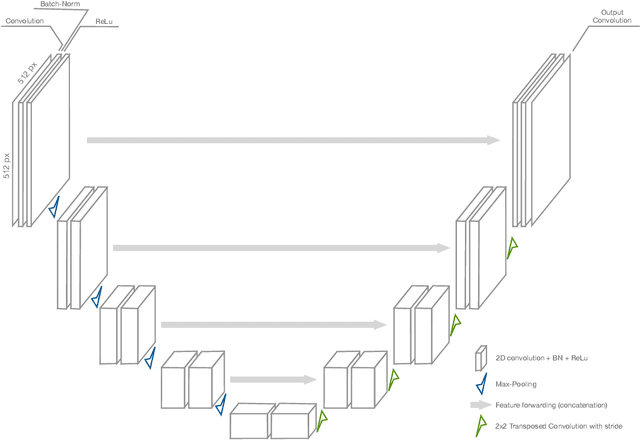
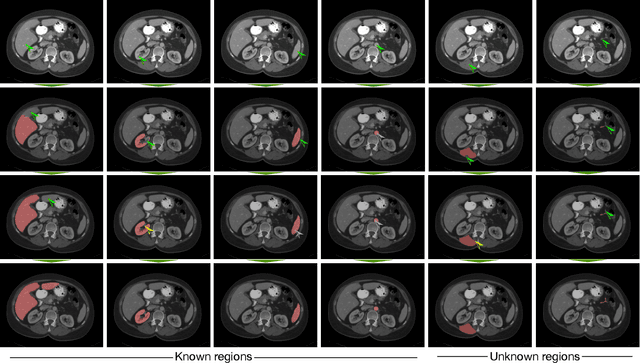
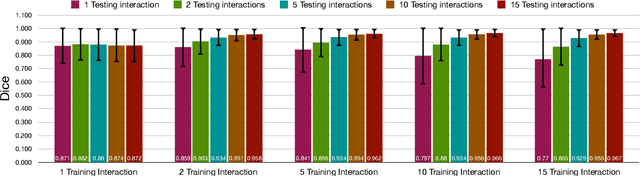
Abstract:Image segmentation plays an essential role in medicine for both diagnostic and interventional tasks. Segmentation approaches are either manual, semi-automated or fully-automated. Manual segmentation offers full control over the quality of the results, but is tedious, time consuming and prone to operator bias. Fully automated methods require no human effort, but often deliver sub-optimal results without providing users with the means to make corrections. Semi-automated approaches keep users in control of the results by providing means for interaction, but the main challenge is to offer a good trade-off between precision and required interaction. In this paper we present a deep learning (DL) based semi-automated segmentation approach that aims to be a "smart" interactive tool for region of interest delineation in medical images. We demonstrate its use for segmenting multiple organs on computed tomography (CT) of the abdomen. Our approach solves some of the most pressing clinical challenges: (i) it requires only one to a few user clicks to deliver excellent 2D segmentations in a fast and reliable fashion; (ii) it can generalize to previously unseen structures and "corner cases"; (iii) it delivers results that can be corrected quickly in a smart and intuitive way up to an arbitrary degree of precision chosen by the user and (iv) ensures high accuracy. We present our approach and compare it to other techniques and previous work to show the advantages brought by our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge