Sean Doyle
Deploying clinical machine learning? Consider the following...
Sep 14, 2021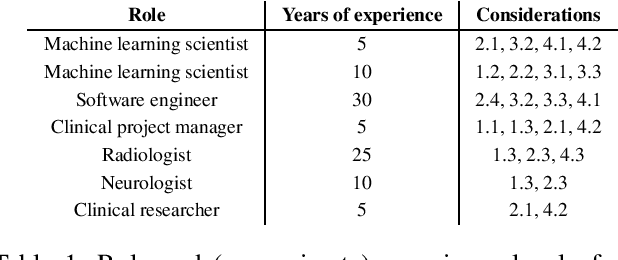
Abstract:Despite the intense attention and investment into clinical machine learning (CML) research, relatively few applications convert to clinical practice. While research is important in advancing the state-of-the-art, translation is equally important in bringing these technologies into a position to ultimately impact patient care and live up to extensive expectations surrounding AI in healthcare. To better characterize a holistic perspective among researchers and practitioners, we survey several participants with experience in developing CML for clinical deployment about their learned experiences. We collate these insights and identify several main categories of barriers and pitfalls in order to better design and develop clinical machine learning applications.
4D CNN for semantic segmentation of cardiac volumetric sequences
Jun 17, 2019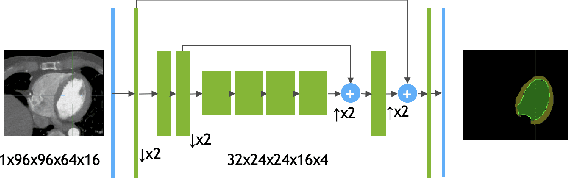
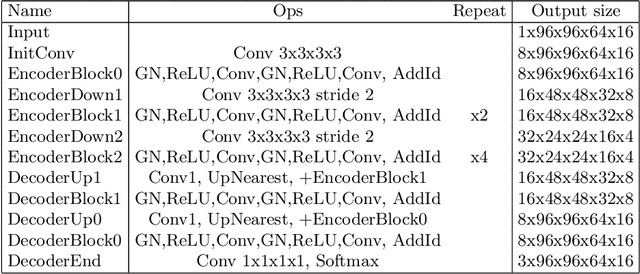
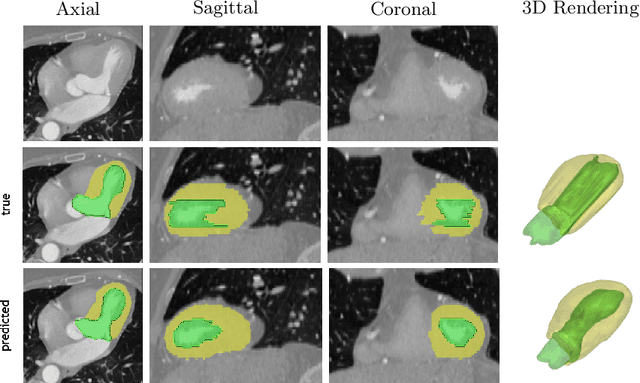
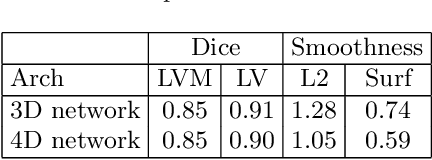
Abstract:We propose a 4D convolutional neural network (CNN) for the segmentation of retrospective ECG-gated cardiac CT, a series of single-channel volumetric data over time. While only a small subset of volumes in the temporal sequence are annotated, we define a sparse loss function on available labels to allow the network to leverage unlabeled images during training and generate a fully segmented sequence. We investigate the accuracy of the proposed 4D network to predict temporally consistent segmentations and compare with traditional 3D segmentation approaches. We demonstrate the feasibility of the 4D CNN and establish its performance on cardiac 4D CCTA.
DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning
Jul 26, 2018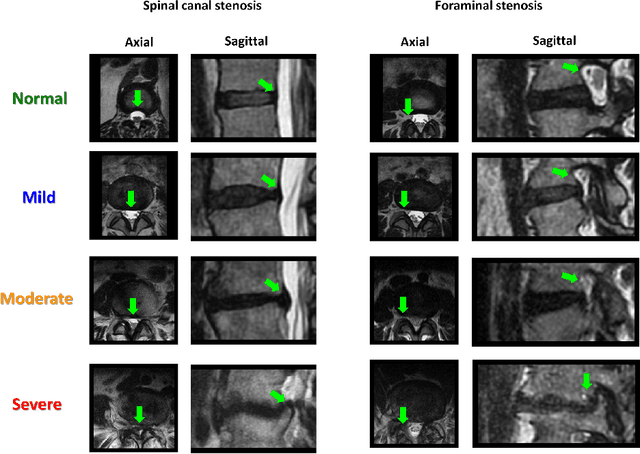
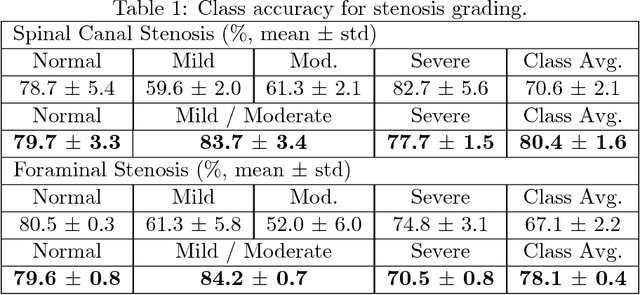
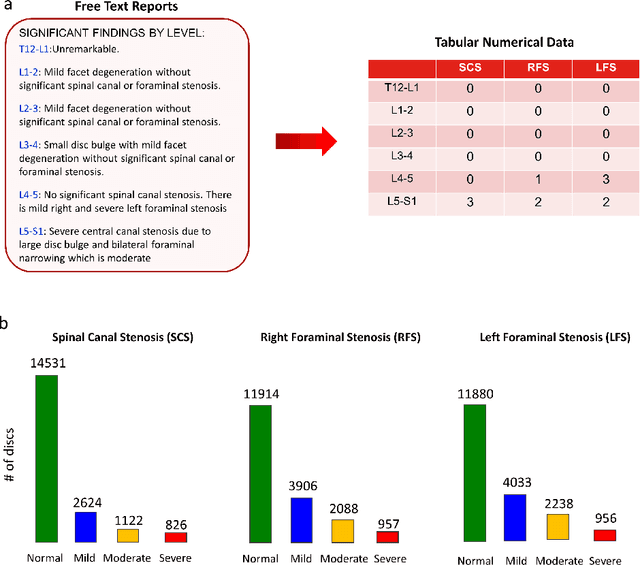
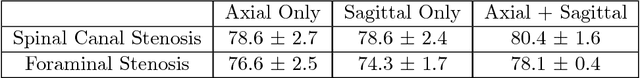
Abstract:The high prevalence of spinal stenosis results in a large volume of MRI imaging, yet interpretation can be time-consuming with high inter-reader variability even among the most specialized radiologists. In this paper, we develop an efficient methodology to leverage the subject-matter-expertise stored in large-scale archival reporting and image data for a deep-learning approach to fully-automated lumbar spinal stenosis grading. Specifically, we introduce three major contributions: (1) a natural-language-processing scheme to extract level-by-level ground-truth labels from free-text radiology reports for the various types and grades of spinal stenosis (2) accurate vertebral segmentation and disc-level localization using a U-Net architecture combined with a spine-curve fitting method, and (3) a multi-input, multi-task, and multi-class convolutional neural network to perform central canal and foraminal stenosis grading on both axial and sagittal imaging series inputs with the extracted report-derived labels applied to corresponding imaging level segments. This study uses a large dataset of 22796 disc-levels extracted from 4075 patients. We achieve state-of-the-art performance on lumbar spinal stenosis classification and expect the technique will increase both radiology workflow efficiency and the perceived value of radiology reports for referring clinicians and patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge