Hanqing Chao
MUSE: Multi-Scale Dense Self-Distillation for Nucleus Detection and Classification
Nov 07, 2025



Abstract:Nucleus detection and classification (NDC) in histopathology analysis is a fundamental task that underpins a wide range of high-level pathology applications. However, existing methods heavily rely on labor-intensive nucleus-level annotations and struggle to fully exploit large-scale unlabeled data for learning discriminative nucleus representations. In this work, we propose MUSE (MUlti-scale denSE self-distillation), a novel self-supervised learning method tailored for NDC. At its core is NuLo (Nucleus-based Local self-distillation), a coordinate-guided mechanism that enables flexible local self-distillation based on predicted nucleus positions. By removing the need for strict spatial alignment between augmented views, NuLo allows critical cross-scale alignment, thus unlocking the capacity of models for fine-grained nucleus-level representation. To support MUSE, we design a simple yet effective encoder-decoder architecture and a large field-of-view semi-supervised fine-tuning strategy that together maximize the value of unlabeled pathology images. Extensive experiments on three widely used benchmarks demonstrate that MUSE effectively addresses the core challenges of histopathological NDC. The resulting models not only surpass state-of-the-art supervised baselines but also outperform generic pathology foundation models.
Neural Proteomics Fields for Super-resolved Spatial Proteomics Prediction
Aug 24, 2025
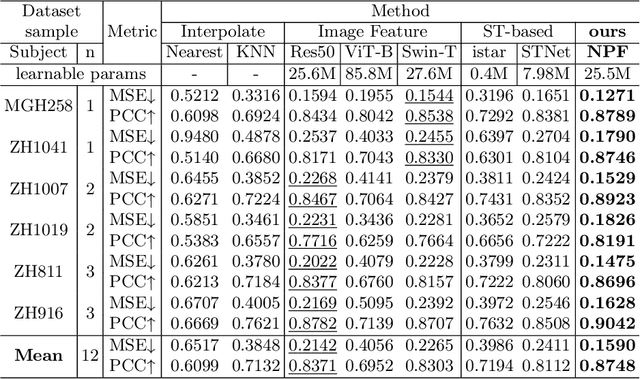

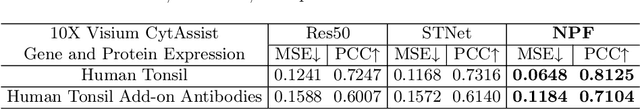
Abstract:Spatial proteomics maps protein distributions in tissues, providing transformative insights for life sciences. However, current sequencing-based technologies suffer from low spatial resolution, and substantial inter-tissue variability in protein expression further compromises the performance of existing molecular data prediction methods. In this work, we introduce the novel task of spatial super-resolution for sequencing-based spatial proteomics (seq-SP) and, to the best of our knowledge, propose the first deep learning model for this task--Neural Proteomics Fields (NPF). NPF formulates seq-SP as a protein reconstruction problem in continuous space by training a dedicated network for each tissue. The model comprises a Spatial Modeling Module, which learns tissue-specific protein spatial distributions, and a Morphology Modeling Module, which extracts tissue-specific morphological features. Furthermore, to facilitate rigorous evaluation, we establish an open-source benchmark dataset, Pseudo-Visium SP, for this task. Experimental results demonstrate that NPF achieves state-of-the-art performance with fewer learnable parameters, underscoring its potential for advancing spatial proteomics research. Our code and dataset are publicly available at https://github.com/Bokai-Zhao/NPF.
From Histopathology Images to Cell Clouds: Learning Slide Representations with Hierarchical Cell Transformer
Dec 21, 2024Abstract:It is clinically crucial and potentially very beneficial to be able to analyze and model directly the spatial distributions of cells in histopathology whole slide images (WSI). However, most existing WSI datasets lack cell-level annotations, owing to the extremely high cost over giga-pixel images. Thus, it remains an open question whether deep learning models can directly and effectively analyze WSIs from the semantic aspect of cell distributions. In this work, we construct a large-scale WSI dataset with more than 5 billion cell-level annotations, termed WSI-Cell5B, and a novel hierarchical Cell Cloud Transformer (CCFormer) to tackle these challenges. WSI-Cell5B is based on 6,998 WSIs of 11 cancers from The Cancer Genome Atlas Program, and all WSIs are annotated per cell by coordinates and types. To the best of our knowledge, WSI-Cell5B is the first WSI-level large-scale dataset integrating cell-level annotations. On the other hand, CCFormer formulates the collection of cells in each WSI as a cell cloud and models cell spatial distribution. Specifically, Neighboring Information Embedding (NIE) is proposed to characterize the distribution of cells within the neighborhood of each cell, and a novel Hierarchical Spatial Perception (HSP) module is proposed to learn the spatial relationship among cells in a bottom-up manner. The clinical analysis indicates that WSI-Cell5B can be used to design clinical evaluation metrics based on counting cells that effectively assess the survival risk of patients. Extensive experiments on survival prediction and cancer staging show that learning from cell spatial distribution alone can already achieve state-of-the-art (SOTA) performance, i.e., CCFormer strongly outperforms other competing methods.
From Pixels to Gigapixels: Bridging Local Inductive Bias and Long-Range Dependencies with Pixel-Mamba
Dec 21, 2024



Abstract:Histopathology plays a critical role in medical diagnostics, with whole slide images (WSIs) offering valuable insights that directly influence clinical decision-making. However, the large size and complexity of WSIs may pose significant challenges for deep learning models, in both computational efficiency and effective representation learning. In this work, we introduce Pixel-Mamba, a novel deep learning architecture designed to efficiently handle gigapixel WSIs. Pixel-Mamba leverages the Mamba module, a state-space model (SSM) with linear memory complexity, and incorporates local inductive biases through progressively expanding tokens, akin to convolutional neural networks. This enables Pixel-Mamba to hierarchically combine both local and global information while efficiently addressing computational challenges. Remarkably, Pixel-Mamba achieves or even surpasses the quantitative performance of state-of-the-art (SOTA) foundation models that were pretrained on millions of WSIs or WSI-text pairs, in a range of tumor staging and survival analysis tasks, {\bf even without requiring any pathology-specific pretraining}. Extensive experiments demonstrate the efficacy of Pixel-Mamba as a powerful and efficient framework for end-to-end WSI analysis.
A Joint Representation Using Continuous and Discrete Features for Cardiovascular Diseases Risk Prediction on Chest CT Scans
Oct 24, 2024



Abstract:Cardiovascular diseases (CVD) remain a leading health concern and contribute significantly to global mortality rates. While clinical advancements have led to a decline in CVD mortality, accurately identifying individuals who could benefit from preventive interventions remains an unsolved challenge in preventive cardiology. Current CVD risk prediction models, recommended by guidelines, are based on limited traditional risk factors or use CT imaging to acquire quantitative biomarkers, and still have limitations in predictive accuracy and applicability. On the other hand, end-to-end trained CVD risk prediction methods leveraging deep learning on CT images often fail to provide transparent and explainable decision grounds for assisting physicians. In this work, we proposed a novel joint representation that integrates discrete quantitative biomarkers and continuous deep features extracted from chest CT scans. Our approach initiated with a deep CVD risk classification model by capturing comprehensive continuous deep learning features while jointly obtaining currently clinical-established quantitative biomarkers via segmentation models. In the feature joint representation stage, we use an instance-wise feature-gated mechanism to align the continuous and discrete features, followed by a soft instance-wise feature interaction mechanism fostering independent and effective feature interaction for the final CVD risk prediction. Our method substantially improves CVD risk predictive performance and offers individual contribution analysis of each biomarker, which is important in assisting physicians' decision-making processes. We validated our method on a public chest low-dose CT dataset and a private external chest standard-dose CT patient cohort of 17,207 CT volumes from 6,393 unique subjects, and demonstrated superior predictive performance, achieving AUCs of 0.875 and 0.843, respectively.
End-to-end Multi-source Visual Prompt Tuning for Survival Analysis in Whole Slide Images
Sep 05, 2024



Abstract:Survival analysis using pathology images poses a considerable challenge, as it requires the localization of relevant information from the multitude of tiles within whole slide images (WSIs). Current methods typically resort to a two-stage approach, where a pre-trained network extracts features from tiles, which are then used by survival models. This process, however, does not optimize the survival models in an end-to-end manner, and the pre-extracted features may not be ideally suited for survival prediction. To address this limitation, we present a novel end-to-end Visual Prompt Tuning framework for survival analysis, named VPTSurv. VPTSurv refines feature embeddings through an efficient encoder-decoder framework. The encoder remains fixed while the framework introduces tunable visual prompts and adaptors, thus permitting end-to-end training specifically for survival prediction by optimizing only the lightweight adaptors and the decoder. Moreover, the versatile VPTSurv framework accommodates multi-source information as prompts, thereby enriching the survival model. VPTSurv achieves substantial increases of 8.7% and 12.5% in the C-index on two immunohistochemical pathology image datasets. These significant improvements highlight the transformative potential of the end-to-end VPT framework over traditional two-stage methods.
Multimodal Neurodegenerative Disease Subtyping Explained by ChatGPT
Jan 31, 2024Abstract:Alzheimer's disease (AD) is the most prevalent neurodegenerative disease; yet its currently available treatments are limited to stopping disease progression. Moreover, effectiveness of these treatments is not guaranteed due to the heterogenetiy of the disease. Therefore, it is essential to be able to identify the disease subtypes at a very early stage. Current data driven approaches are able to classify the subtypes at later stages of AD or related disorders, but struggle when predicting at the asymptomatic or prodromal stage. Moreover, most existing models either lack explainability behind the classification or only use a single modality for the assessment, limiting scope of its analysis. Thus, we propose a multimodal framework that uses early-stage indicators such as imaging, genetics and clinical assessments to classify AD patients into subtypes at early stages. Similarly, we build prompts and use large language models, such as ChatGPT, to interpret the findings of our model. In our framework, we propose a tri-modal co-attention mechanism (Tri-COAT) to explicitly learn the cross-modal feature associations. Our proposed model outperforms baseline models and provides insight into key cross-modal feature associations supported by known biological mechanisms.
Spectral Adversarial MixUp for Few-Shot Unsupervised Domain Adaptation
Sep 03, 2023Abstract:Domain shift is a common problem in clinical applications, where the training images (source domain) and the test images (target domain) are under different distributions. Unsupervised Domain Adaptation (UDA) techniques have been proposed to adapt models trained in the source domain to the target domain. However, those methods require a large number of images from the target domain for model training. In this paper, we propose a novel method for Few-Shot Unsupervised Domain Adaptation (FSUDA), where only a limited number of unlabeled target domain samples are available for training. To accomplish this challenging task, first, a spectral sensitivity map is introduced to characterize the generalization weaknesses of models in the frequency domain. We then developed a Sensitivity-guided Spectral Adversarial MixUp (SAMix) method to generate target-style images to effectively suppresses the model sensitivity, which leads to improved model generalizability in the target domain. We demonstrated the proposed method and rigorously evaluated its performance on multiple tasks using several public datasets.
When Neural Networks Fail to Generalize? A Model Sensitivity Perspective
Dec 01, 2022Abstract:Domain generalization (DG) aims to train a model to perform well in unseen domains under different distributions. This paper considers a more realistic yet more challenging scenario,namely Single Domain Generalization (Single-DG), where only a single source domain is available for training. To tackle this challenge, we first try to understand when neural networks fail to generalize? We empirically ascertain a property of a model that correlates strongly with its generalization that we coin as "model sensitivity". Based on our analysis, we propose a novel strategy of Spectral Adversarial Data Augmentation (SADA) to generate augmented images targeted at the highly sensitive frequencies. Models trained with these hard-to-learn samples can effectively suppress the sensitivity in the frequency space, which leads to improved generalization performance. Extensive experiments on multiple public datasets demonstrate the superiority of our approach, which surpasses the state-of-the-art single-DG methods.
Regression Metric Loss: Learning a Semantic Representation Space for Medical Images
Jul 12, 2022
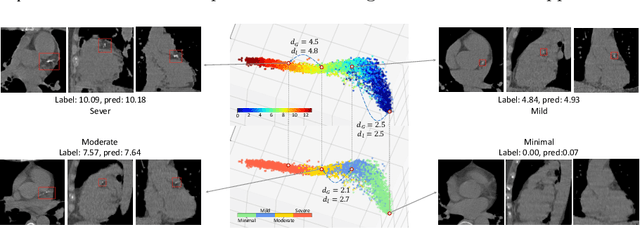
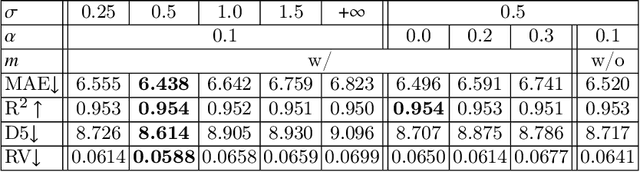
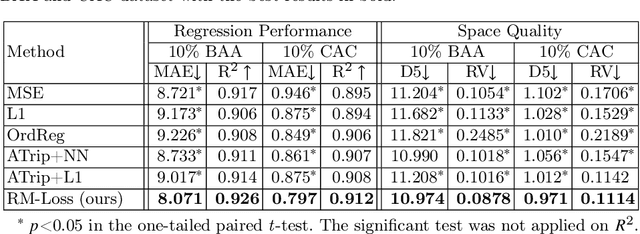
Abstract:Regression plays an essential role in many medical imaging applications for estimating various clinical risk or measurement scores. While training strategies and loss functions have been studied for the deep neural networks in medical image classification tasks, options for regression tasks are very limited. One of the key challenges is that the high-dimensional feature representation learned by existing popular loss functions like Mean Squared Error or L1 loss is hard to interpret. In this paper, we propose a novel Regression Metric Loss (RM-Loss), which endows the representation space with the semantic meaning of the label space by finding a representation manifold that is isometric to the label space. Experiments on two regression tasks, i.e. coronary artery calcium score estimation and bone age assessment, show that RM-Loss is superior to the existing popular regression losses on both performance and interpretability. Code is available at https://github.com/DIAL-RPI/Regression-Metric-Loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge