Geert Litjens
The Medical Segmentation Decathlon
Jun 10, 2021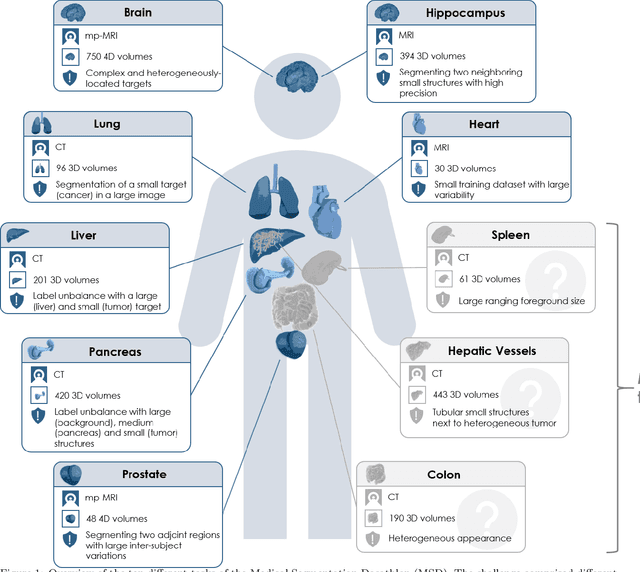
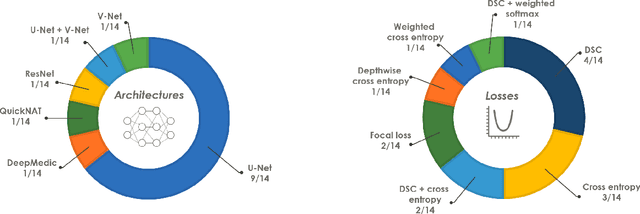
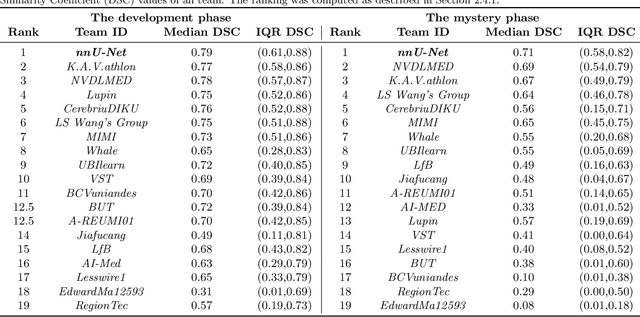
Abstract:International challenges have become the de facto standard for comparative assessment of image analysis algorithms given a specific task. Segmentation is so far the most widely investigated medical image processing task, but the various segmentation challenges have typically been organized in isolation, such that algorithm development was driven by the need to tackle a single specific clinical problem. We hypothesized that a method capable of performing well on multiple tasks will generalize well to a previously unseen task and potentially outperform a custom-designed solution. To investigate the hypothesis, we organized the Medical Segmentation Decathlon (MSD) - a biomedical image analysis challenge, in which algorithms compete in a multitude of both tasks and modalities. The underlying data set was designed to explore the axis of difficulties typically encountered when dealing with medical images, such as small data sets, unbalanced labels, multi-site data and small objects. The MSD challenge confirmed that algorithms with a consistent good performance on a set of tasks preserved their good average performance on a different set of previously unseen tasks. Moreover, by monitoring the MSD winner for two years, we found that this algorithm continued generalizing well to a wide range of other clinical problems, further confirming our hypothesis. Three main conclusions can be drawn from this study: (1) state-of-the-art image segmentation algorithms are mature, accurate, and generalize well when retrained on unseen tasks; (2) consistent algorithmic performance across multiple tasks is a strong surrogate of algorithmic generalizability; (3) the training of accurate AI segmentation models is now commoditized to non AI experts.
Common Limitations of Image Processing Metrics: A Picture Story
Apr 13, 2021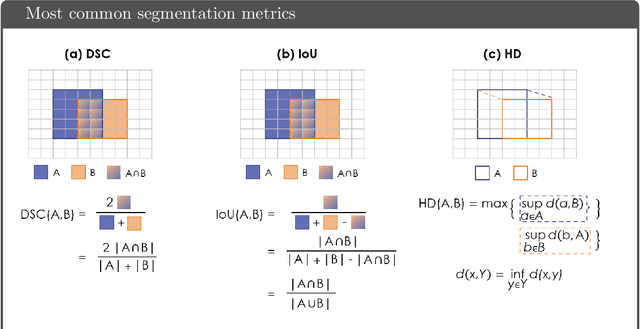
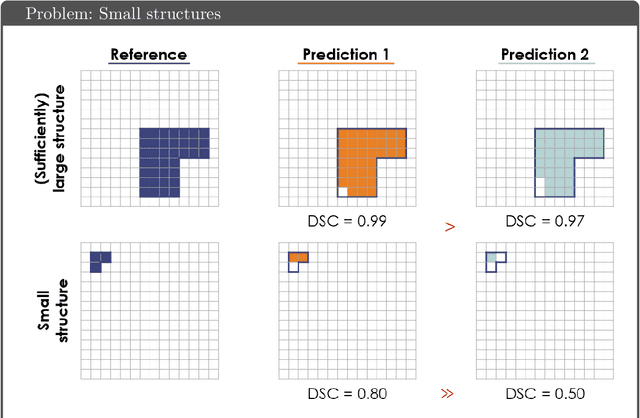
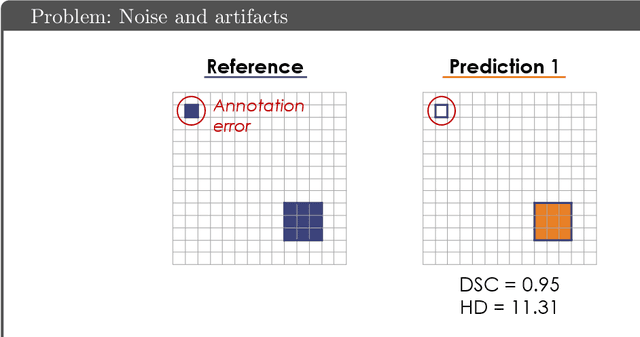
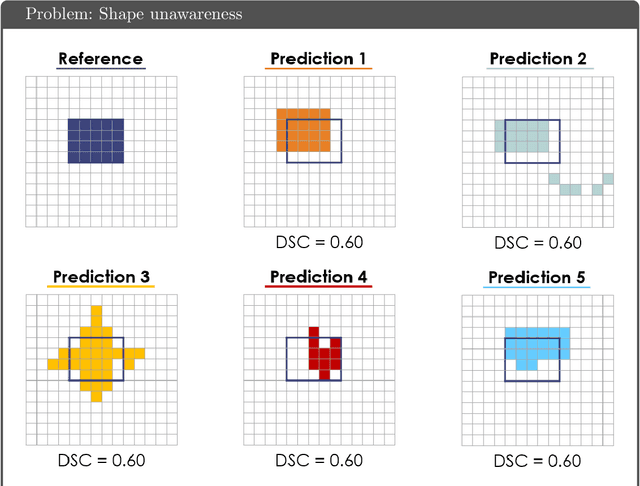
Abstract:While the importance of automatic image analysis is increasing at an enormous pace, recent meta-research revealed major flaws with respect to algorithm validation. Specifically, performance metrics are key for objective, transparent and comparative performance assessment, but relatively little attention has been given to the practical pitfalls when using specific metrics for a given image analysis task. A common mission of several international initiatives is therefore to provide researchers with guidelines and tools to choose the performance metrics in a problem-aware manner. This dynamically updated document has the purpose to illustrate important limitations of performance metrics commonly applied in the field of image analysis. The current version is based on a Delphi process on metrics conducted by an international consortium of image analysis experts.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020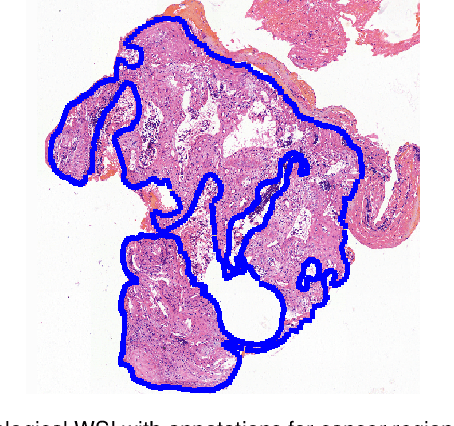
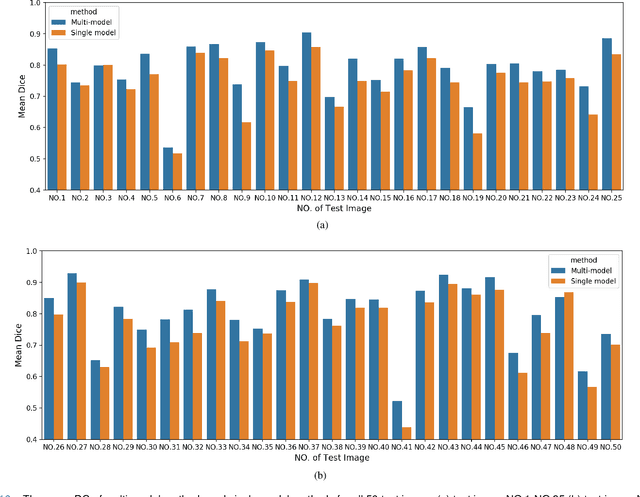
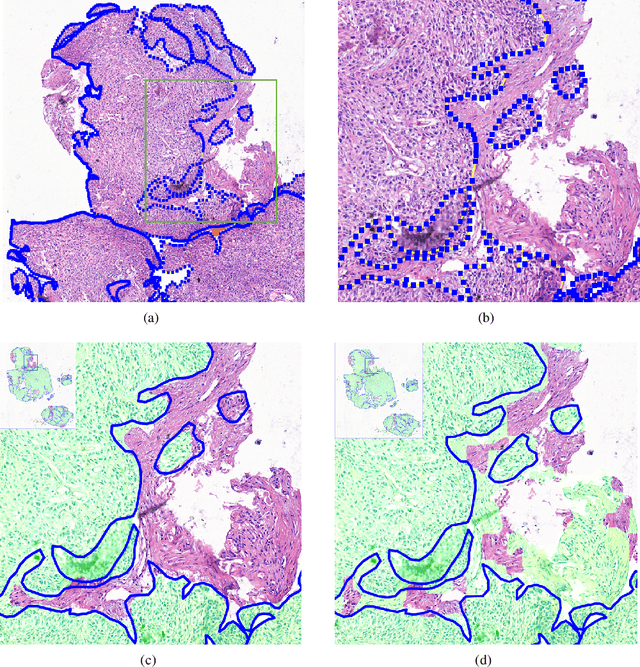
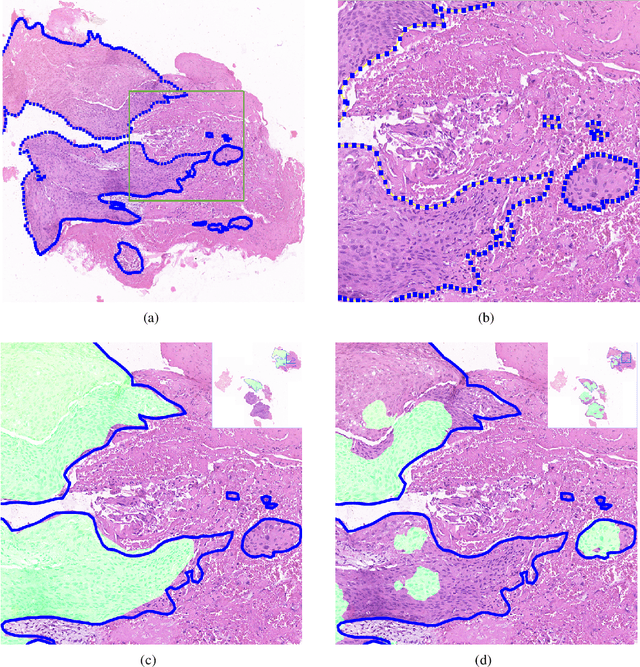
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels
Jun 05, 2020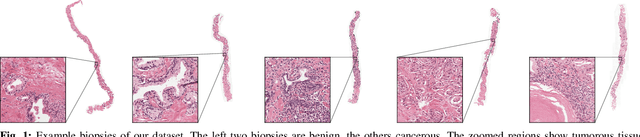
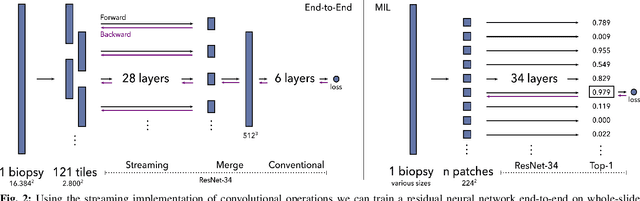
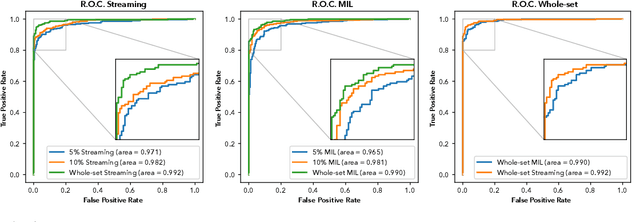

Abstract:Prostate cancer is the most prevalent cancer among men in Western countries, with 1.1 million new diagnoses every year. The gold standard for the diagnosis of prostate cancer is a pathologists' evaluation of prostate tissue. To potentially assist pathologists deep-learning-based cancer detection systems have been developed. Many of the state-of-the-art models are patch-based convolutional neural networks, as the use of entire scanned slides is hampered by memory limitations on accelerator cards. Patch-based systems typically require detailed, pixel-level annotations for effective training. However, such annotations are seldom readily available, in contrast to the clinical reports of pathologists, which contain slide-level labels. As such, developing algorithms which do not require manual pixel-wise annotations, but can learn using only the clinical report would be a significant advancement for the field. In this paper, we propose to use a streaming implementation of convolutional layers, to train a modern CNN (ResNet-34) with 21 million parameters end-to-end on 4712 prostate biopsies. The method enables the use of entire biopsy images at high-resolution directly by reducing the GPU memory requirements by 2.4 TB. We show that modern CNNs, trained using our streaming approach, can extract meaningful features from high-resolution images without additional heuristics, reaching similar performance as state-of-the-art patch-based and multiple-instance learning methods. By circumventing the need for manual annotations, this approach can function as a blueprint for other tasks in histopathological diagnosis. The source code to reproduce the streaming models is available at https://github.com/DIAGNijmegen/pathology-streaming-pipeline .
Artificial Intelligence Assistance Significantly Improves Gleason Grading of Prostate Biopsies by Pathologists
Feb 11, 2020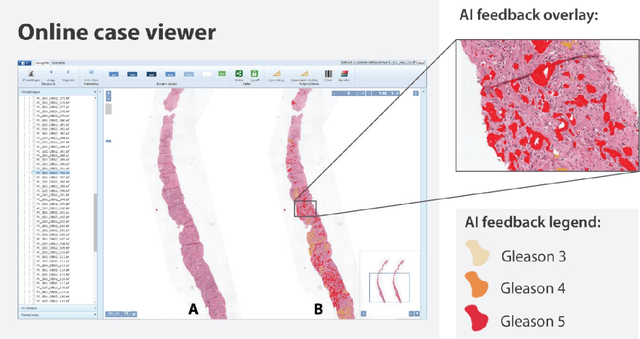
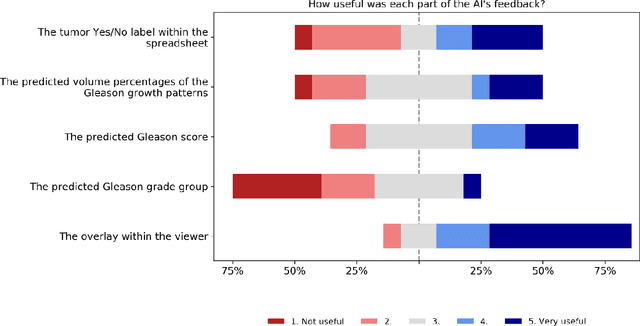
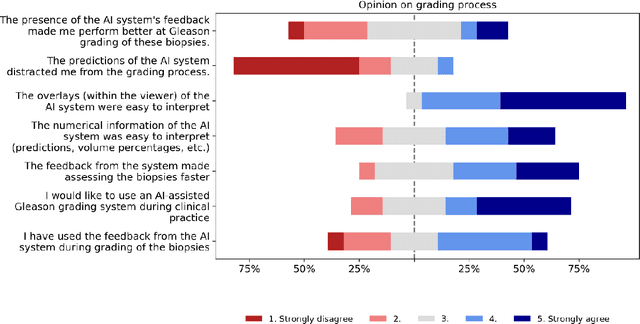
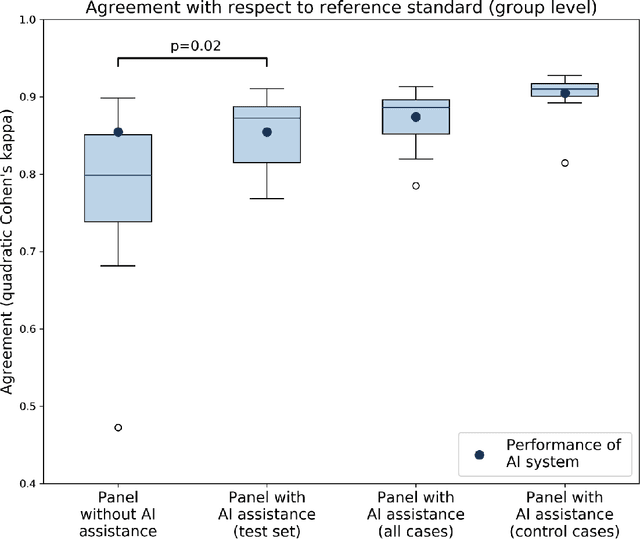
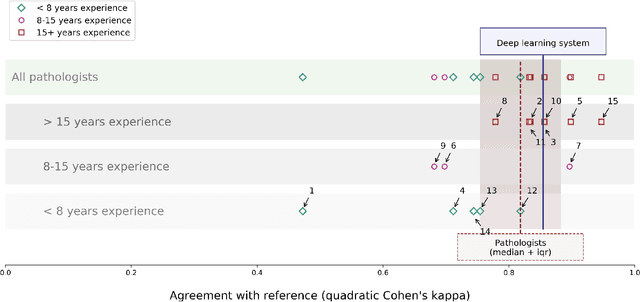
Abstract:While the Gleason score is the most important prognostic marker for prostate cancer patients, it suffers from significant observer variability. Artificial Intelligence (AI) systems, based on deep learning, have proven to achieve pathologist-level performance at Gleason grading. However, the performance of such systems can degrade in the presence of artifacts, foreign tissue, or other anomalies. Pathologists integrating their expertise with feedback from an AI system could result in a synergy that outperforms both the individual pathologist and the system. Despite the hype around AI assistance, existing literature on this topic within the pathology domain is limited. We investigated the value of AI assistance for grading prostate biopsies. A panel of fourteen observers graded 160 biopsies with and without AI assistance. Using AI, the agreement of the panel with an expert reference standard significantly increased (quadratically weighted Cohen's kappa, 0.799 vs 0.872; p=0.018). Our results show the added value of AI systems for Gleason grading, but more importantly, show the benefits of pathologist-AI synergy.
Streaming convolutional neural networks for end-to-end learning with multi-megapixel images
Nov 11, 2019



Abstract:Due to memory constraints on current hardware, most convolution neural networks (CNN) are trained on sub-megapixel images. For example, most popular datasets in computer vision contain images much less than a megapixel in size (0.09MP for ImageNet and 0.001MP for CIFAR-10). In some domains such as medical imaging, multi-megapixel images are needed to identify the presence of disease accurately. We propose a novel method to directly train convolutional neural networks using any input image size end-to-end. This method exploits the locality of most operations in modern convolutional neural networks by performing the forward and backward pass on smaller tiles of the image. In this work, we show a proof of concept using images of up to 66-megapixels (8192x8192), saving approximately 50GB of memory per image. Using two public challenge datasets, we demonstrate that CNNs can learn to extract relevant information from these large images and benefit from increasing resolution. We improved the area under the receiver-operating characteristic curve from 0.580 (4MP) to 0.706 (66MP) for metastasis detection in breast cancer (CAMELYON17). We also obtained a Spearman correlation metric approaching state-of-the-art performance on the TUPAC16 dataset, from 0.485 (1MP) to 0.570 (16MP). Code to reproduce a subset of the experiments is available at https://github.com/DIAGNijmegen/StreamingCNN.
Neural Ordinary Differential Equations for Semantic Segmentation of Individual Colon Glands
Oct 23, 2019

Abstract:Automated medical image segmentation plays a key role in quantitative research and diagnostics. Convolutional neural networks based on the U-Net architecture are the state-of-the-art. A key disadvantage is the hard-coding of the receptive field size, which requires architecture optimization for each segmentation task. Furthermore, increasing the receptive field results in an increasing number of weights. Recently, Neural Ordinary Differential Equations (NODE) have been proposed, a new type of continuous depth deep neural network. This framework allows for a dynamic receptive field at a fixed memory cost and a smaller amount of parameters. We show on a colon gland segmentation dataset (GlaS) that these NODEs can be used within the U-Net framework to improve segmentation results while reducing memory load and parameter counts.
Automated Gleason Grading of Prostate Biopsies using Deep Learning
Jul 18, 2019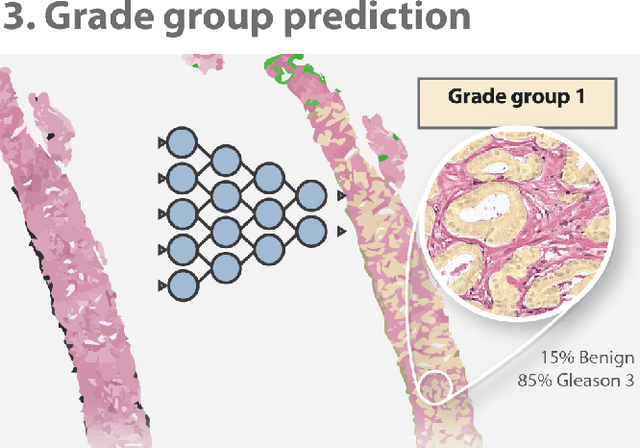
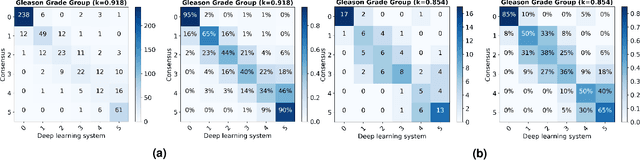
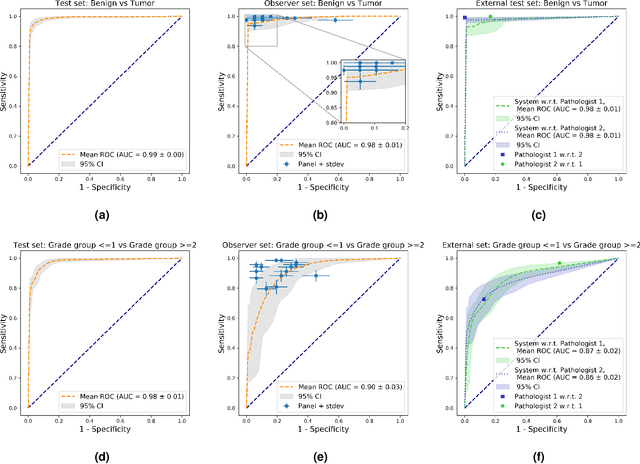
Abstract:The Gleason score is the most important prognostic marker for prostate cancer patients but suffers from significant inter-observer variability. We developed a fully automated deep learning system to grade prostate biopsies. The system was developed using 5834 biopsies from 1243 patients. A semi-automatic labeling technique was used to circumvent the need for full manual annotation by pathologists. The developed system achieved a high agreement with the reference standard. In a separate observer experiment, the deep learning system outperformed 10 out of 15 pathologists. The system has the potential to improve prostate cancer prognostics by acting as a first or second reader.
Dealing with Label Scarcity in Computational Pathology: A Use Case in Prostate Cancer Classification
May 16, 2019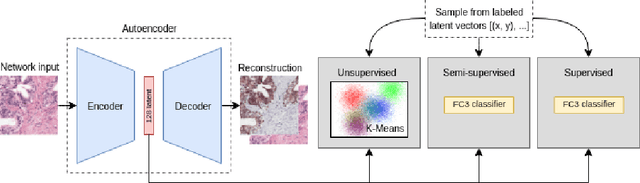
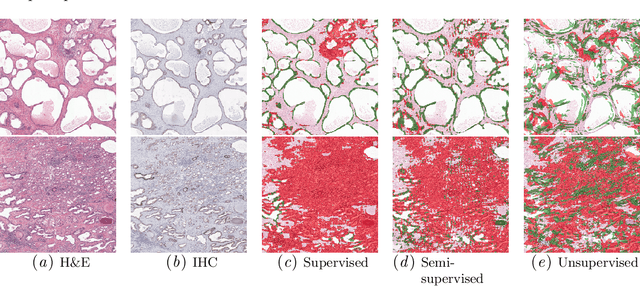
Abstract:Large amounts of unlabelled data are commonplace for many applications in computational pathology, whereas labelled data is often expensive, both in time and cost, to acquire. We investigate the performance of unsupervised and supervised deep learning methods when few labelled data are available. Three methods are compared: clustering autoencoder latent vectors (unsupervised), a single layer classifier combined with a pre-trained autoencoder (semi-supervised), and a supervised CNN. We apply these methods on hematoxylin and eosin (H&E) stained prostatectomy images to classify tumour versus non-tumour tissue. Results show that semi-/unsupervised methods have an advantage over supervised learning when few labels are available. Additionally, we show that incorporating immunohistochemistry (IHC) stained data provides an increase in performance over only using H&E.
A large annotated medical image dataset for the development and evaluation of segmentation algorithms
Feb 25, 2019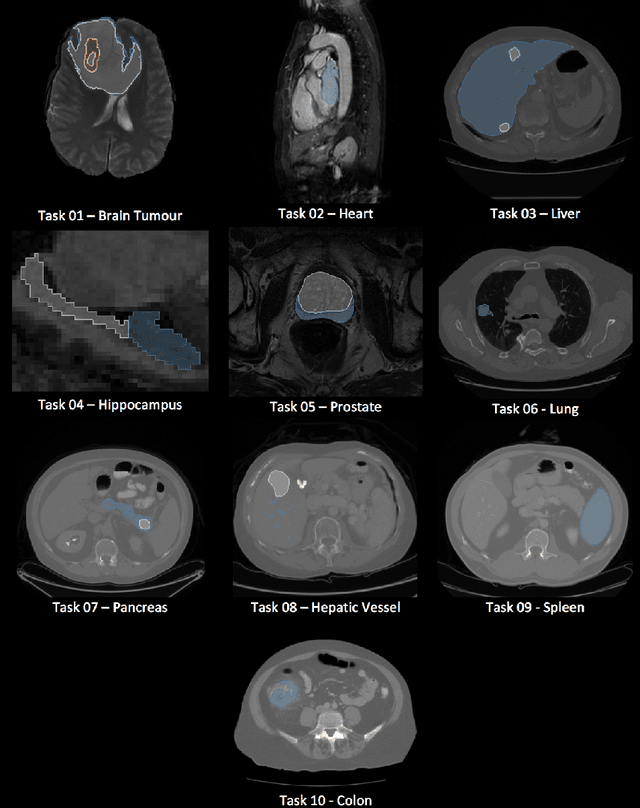

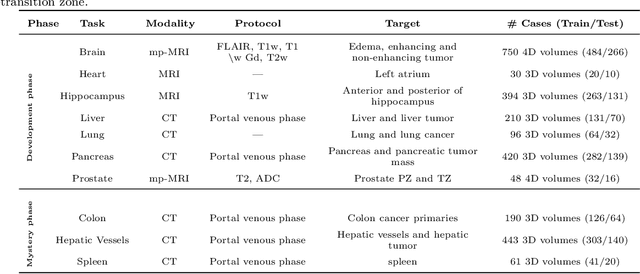
Abstract:Semantic segmentation of medical images aims to associate a pixel with a label in a medical image without human initialization. The success of semantic segmentation algorithms is contingent on the availability of high-quality imaging data with corresponding labels provided by experts. We sought to create a large collection of annotated medical image datasets of various clinically relevant anatomies available under open source license to facilitate the development of semantic segmentation algorithms. Such a resource would allow: 1) objective assessment of general-purpose segmentation methods through comprehensive benchmarking and 2) open and free access to medical image data for any researcher interested in the problem domain. Through a multi-institutional effort, we generated a large, curated dataset representative of several highly variable segmentation tasks that was used in a crowd-sourced challenge - the Medical Segmentation Decathlon held during the 2018 Medical Image Computing and Computer Aided Interventions Conference in Granada, Spain. Here, we describe these ten labeled image datasets so that these data may be effectively reused by the research community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge