Jeroen van der Laak
Democratizing Pathology Co-Pilots: An Open Pipeline and Dataset for Whole-Slide Vision-Language Modelling
Dec 19, 2025



Abstract:Vision-language models (VLMs) have the potential to become co-pilots for pathologists. However, most VLMs either focus on small regions of interest within whole-slide images, provide only static slide-level outputs, or rely on data that is not publicly available, limiting reproducibility. Furthermore, training data containing WSIs paired with detailed clinical reports is scarce, restricting progress toward transparent and generalisable VLMs. We address these limitations with three main contributions. First, we introduce Polysome, a standardised tool for synthetic instruction generation. Second, we apply Polysome to the public HISTAI dataset, generating HISTAI-Instruct, a large whole-slide instruction tuning dataset spanning 24,259 slides and over 1.1 million instruction-response pairs. Finally, we use HISTAI-Instruct to train ANTONI-α, a VLM capable of visual-question answering (VQA). We show that ANTONI-α outperforms MedGemma on WSI-level VQA tasks of tissue identification, neoplasm detection, and differential diagnosis. We also compare the performance of multiple incarnations of ANTONI-α trained with different amounts of data. All methods, data, and code are publicly available.
A Multicentric Dataset for Training and Benchmarking Breast Cancer Segmentation in H&E Slides
Oct 02, 2025


Abstract:Automated semantic segmentation of whole-slide images (WSIs) stained with hematoxylin and eosin (H&E) is essential for large-scale artificial intelligence-based biomarker analysis in breast cancer. However, existing public datasets for breast cancer segmentation lack the morphological diversity needed to support model generalizability and robust biomarker validation across heterogeneous patient cohorts. We introduce BrEast cancEr hisTopathoLogy sEgmentation (BEETLE), a dataset for multiclass semantic segmentation of H&E-stained breast cancer WSIs. It consists of 587 biopsies and resections from three collaborating clinical centers and two public datasets, digitized using seven scanners, and covers all molecular subtypes and histological grades. Using diverse annotation strategies, we collected annotations across four classes - invasive epithelium, non-invasive epithelium, necrosis, and other - with particular focus on morphologies underrepresented in existing datasets, such as ductal carcinoma in situ and dispersed lobular tumor cells. The dataset's diversity and relevance to the rapidly growing field of automated biomarker quantification in breast cancer ensure its high potential for reuse. Finally, we provide a well-curated, multicentric external evaluation set to enable standardized benchmarking of breast cancer segmentation models.
"No negatives needed": weakly-supervised regression for interpretable tumor detection in whole-slide histopathology images
Feb 28, 2025



Abstract:Accurate tumor detection in digital pathology whole-slide images (WSIs) is crucial for cancer diagnosis and treatment planning. Multiple Instance Learning (MIL) has emerged as a widely used approach for weakly-supervised tumor detection with large-scale data without the need for manual annotations. However, traditional MIL methods often depend on classification tasks that require tumor-free cases as negative examples, which are challenging to obtain in real-world clinical workflows, especially for surgical resection specimens. We address this limitation by reformulating tumor detection as a regression task, estimating tumor percentages from WSIs, a clinically available target across multiple cancer types. In this paper, we provide an analysis of the proposed weakly-supervised regression framework by applying it to multiple organs, specimen types and clinical scenarios. We characterize the robustness of our framework to tumor percentage as a noisy regression target, and introduce a novel concept of amplification technique to improve tumor detection sensitivity when learning from small tumor regions. Finally, we provide interpretable insights into the model's predictions by analyzing visual attention and logit maps. Our code is available at https://github.com/DIAGNijmegen/tumor-percentage-mil-regression.
Improving Quality Control of Whole Slide Images by Explicit Artifact Augmentation
Jun 17, 2024Abstract:The problem of artifacts in whole slide image acquisition, prevalent in both clinical workflows and research-oriented settings, necessitates human intervention and re-scanning. Overcoming this challenge requires developing quality control algorithms, that are hindered by the limited availability of relevant annotated data in histopathology. The manual annotation of ground-truth for artifact detection methods is expensive and time-consuming. This work addresses the issue by proposing a method dedicated to augmenting whole slide images with artifacts. The tool seamlessly generates and blends artifacts from an external library to a given histopathology dataset. The augmented datasets are then utilized to train artifact classification methods. The evaluation shows their usefulness in classification of the artifacts, where they show an improvement from 0.10 to 0.01 AUROC depending on the artifact type. The framework, model, weights, and ground-truth annotations are freely released to facilitate open science and reproducible research.
Masked Attention as a Mechanism for Improving Interpretability of Vision Transformers
Apr 28, 2024



Abstract:Vision Transformers are at the heart of the current surge of interest in foundation models for histopathology. They process images by breaking them into smaller patches following a regular grid, regardless of their content. Yet, not all parts of an image are equally relevant for its understanding. This is particularly true in computational pathology where background is completely non-informative and may introduce artefacts that could mislead predictions. To address this issue, we propose a novel method that explicitly masks background in Vision Transformers' attention mechanism. This ensures tokens corresponding to background patches do not contribute to the final image representation, thereby improving model robustness and interpretability. We validate our approach using prostate cancer grading from whole-slide images as a case study. Our results demonstrate that it achieves comparable performance with plain self-attention while providing more accurate and clinically meaningful attention heatmaps.
Hierarchical Vision Transformers for Context-Aware Prostate Cancer Grading in Whole Slide Images
Dec 19, 2023Abstract:Vision Transformers (ViTs) have ushered in a new era in computer vision, showcasing unparalleled performance in many challenging tasks. However, their practical deployment in computational pathology has largely been constrained by the sheer size of whole slide images (WSIs), which result in lengthy input sequences. Transformers faced a similar limitation when applied to long documents, and Hierarchical Transformers were introduced to circumvent it. Given the analogous challenge with WSIs and their inherent hierarchical structure, Hierarchical Vision Transformers (H-ViTs) emerge as a promising solution in computational pathology. This work delves into the capabilities of H-ViTs, evaluating their efficiency for prostate cancer grading in WSIs. Our results show that they achieve competitive performance against existing state-of-the-art solutions.
LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset
Jan 16, 2023



Abstract:We introduce LYSTO, the Lymphocyte Assessment Hackathon, which was held in conjunction with the MICCAI 2019 Conference in Shenzen (China). The competition required participants to automatically assess the number of lymphocytes, in particular T-cells, in histopathological images of colon, breast, and prostate cancer stained with CD3 and CD8 immunohistochemistry. Differently from other challenges setup in medical image analysis, LYSTO participants were solely given a few hours to address this problem. In this paper, we describe the goal and the multi-phase organization of the hackathon; we describe the proposed methods and the on-site results. Additionally, we present post-competition results where we show how the presented methods perform on an independent set of lung cancer slides, which was not part of the initial competition, as well as a comparison on lymphocyte assessment between presented methods and a panel of pathologists. We show that some of the participants were capable to achieve pathologist-level performance at lymphocyte assessment. After the hackathon, LYSTO was left as a lightweight plug-and-play benchmark dataset on grand-challenge website, together with an automatic evaluation platform. LYSTO has supported a number of research in lymphocyte assessment in oncology. LYSTO will be a long-lasting educational challenge for deep learning and digital pathology, it is available at https://lysto.grand-challenge.org/.
Domain adaptation strategies for cancer-independent detection of lymph node metastases
Jul 13, 2022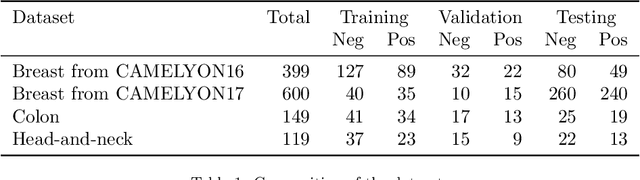
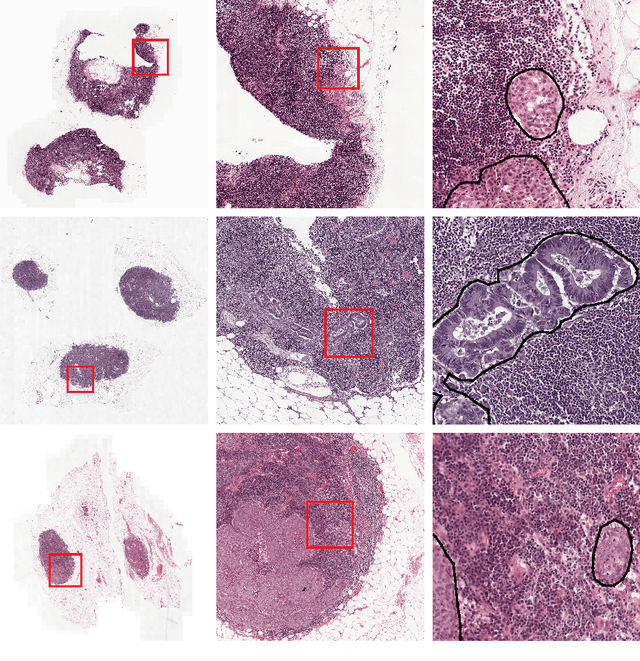
Abstract:Recently, large, high-quality public datasets have led to the development of convolutional neural networks that can detect lymph node metastases of breast cancer at the level of expert pathologists. Many cancers, regardless of the site of origin, can metastasize to lymph nodes. However, collecting and annotating high-volume, high-quality datasets for every cancer type is challenging. In this paper we investigate how to leverage existing high-quality datasets most efficiently in multi-task settings for closely related tasks. Specifically, we will explore different training and domain adaptation strategies, including prevention of catastrophic forgetting, for colon and head-and-neck cancer metastasis detection in lymph nodes. Our results show state-of-the-art performance on both cancer metastasis detection tasks. Furthermore, we show the effectiveness of repeated adaptation of networks from one cancer type to another to obtain multi-task metastasis detection networks. Last, we show that leveraging existing high-quality datasets can significantly boost performance on new target tasks and that catastrophic forgetting can be effectively mitigated using regularization.
Automated risk classification of colon biopsies based on semantic segmentation of histopathology images
Sep 16, 2021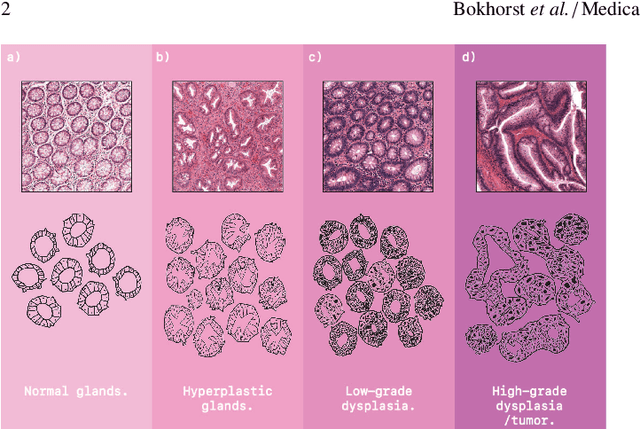
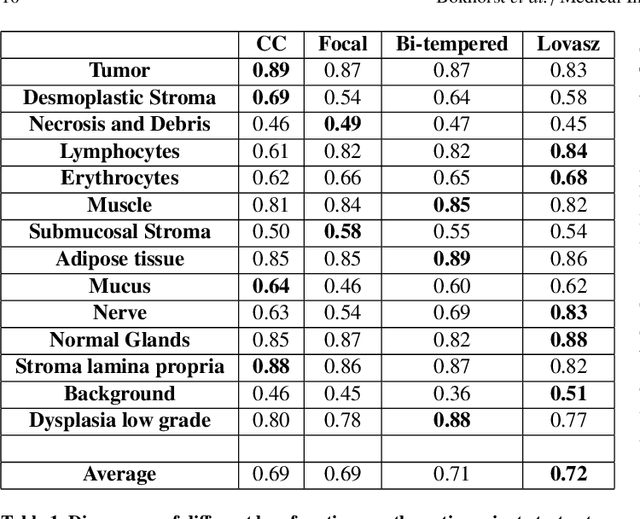
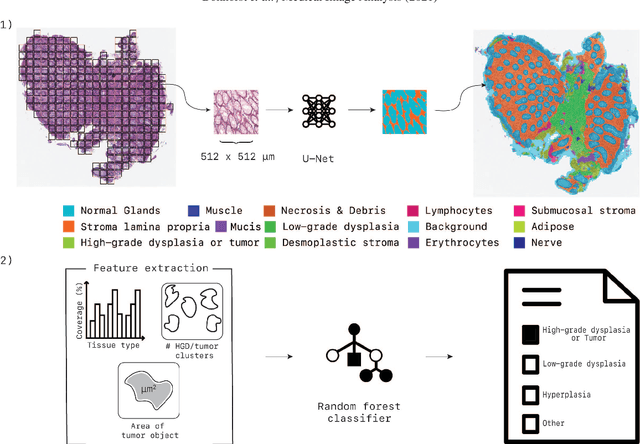
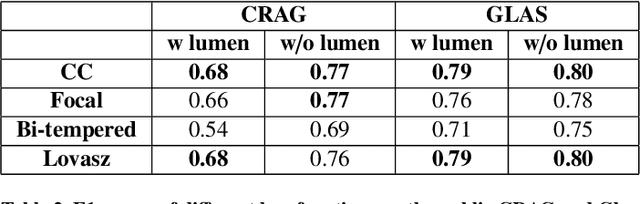
Abstract:Artificial Intelligence (AI) can potentially support histopathologists in the diagnosis of a broad spectrum of cancer types. In colorectal cancer (CRC), AI can alleviate the laborious task of characterization and reporting on resected biopsies, including polyps, the numbers of which are increasing as a result of CRC population screening programs, ongoing in many countries all around the globe. Here, we present an approach to address two major challenges in automated assessment of CRC histopathology whole-slide images. First, we present an AI-based method to segment multiple tissue compartments in the H\&E-stained whole-slide image, which provides a different, more perceptible picture of tissue morphology and composition. We test and compare a panel of state-of-the-art loss functions available for segmentation models, and provide indications about their use in histopathology image segmentation, based on the analysis of a) a multi-centric cohort of CRC cases from five medical centers in the Netherlands and Germany, and b) two publicly available datasets on segmentation in CRC. Second, we use the best performing AI model as the basis for a computer-aided diagnosis system (CAD) that classifies colon biopsies into four main categories that are relevant pathologically. We report the performance of this system on an independent cohort of more than 1,000 patients. The results show the potential of such an AI-based system to assist pathologists in diagnosis of CRC in the context of population screening. We have made the segmentation model available for research use on https://grand-challenge.org/algorithms/colon-tissue-segmentation/.
High-resolution Image Registration of Consecutive and Re-stained Sections in Histopathology
Jun 24, 2021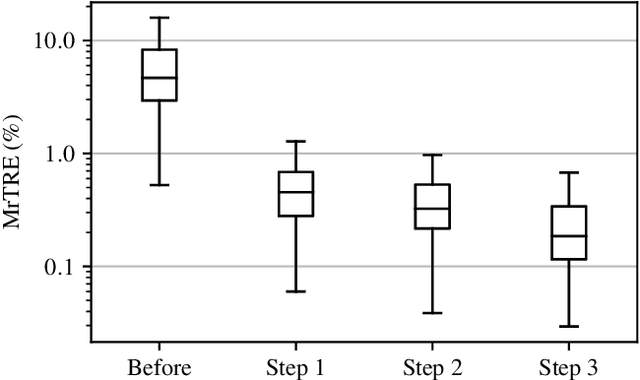
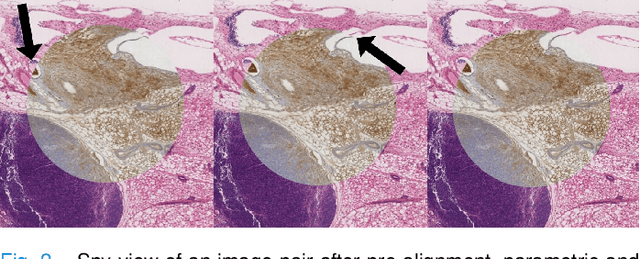
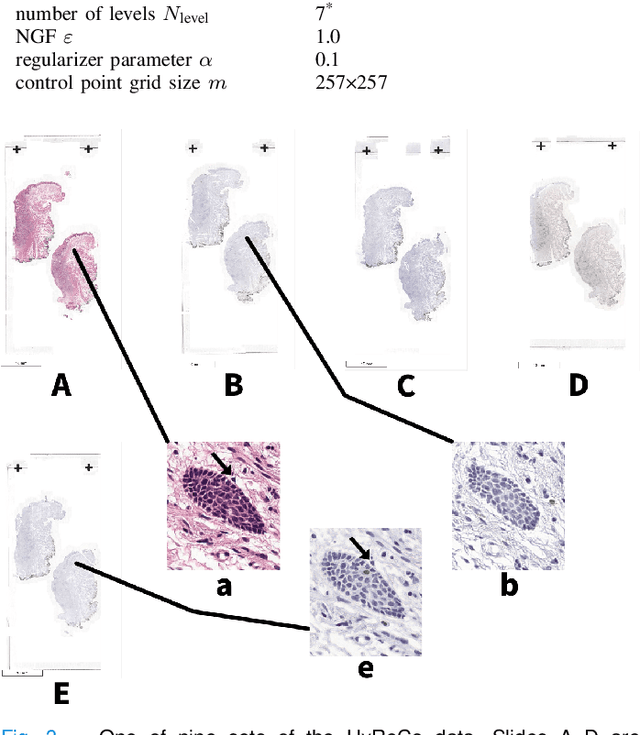
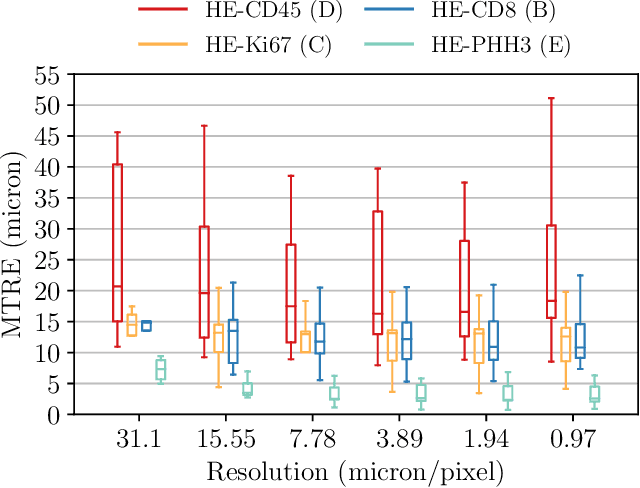
Abstract:We compare variational image registration in consectutive and re-stained sections from histopathology. We present a fully-automatic algorithm for non-parametric (nonlinear) image registration and apply it to a previously existing dataset from the ANHIR challenge (230 slide pairs, consecutive sections) and a new dataset (hybrid re-stained and consecutive, 81 slide pairs, ca. 3000 landmarks) which is made publicly available. Registration hyperparameters are obtained in the ANHIR dataset and applied to the new dataset without modification. In the new dataset, landmark errors after registration range from 13.2 micrometers for consecutive sections to 1 micrometer for re-stained sections. We observe that non-parametric registration leads to lower landmark errors in both cases, even though the effect is smaller in re-stained sections. The nucleus-level alignment after non-parametric registration of re-stained sections provides a valuable tool to generate automatic ground-truth for machine learning applications in histopathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge