Eli Gibson
Revisiting 2D Foundation Models for Scalable 3D Medical Image Classification
Dec 15, 2025



Abstract:3D medical image classification is essential for modern clinical workflows. Medical foundation models (FMs) have emerged as a promising approach for scaling to new tasks, yet current research suffers from three critical pitfalls: data-regime bias, suboptimal adaptation, and insufficient task coverage. In this paper, we address these pitfalls and introduce AnyMC3D, a scalable 3D classifier adapted from 2D FMs. Our method scales efficiently to new tasks by adding only lightweight plugins (about 1M parameters per task) on top of a single frozen backbone. This versatile framework also supports multi-view inputs, auxiliary pixel-level supervision, and interpretable heatmap generation. We establish a comprehensive benchmark of 12 tasks covering diverse pathologies, anatomies, and modalities, and systematically analyze state-of-the-art 3D classification techniques. Our analysis reveals key insights: (1) effective adaptation is essential to unlock FM potential, (2) general-purpose FMs can match medical-specific FMs if properly adapted, and (3) 2D-based methods surpass 3D architectures for 3D classification. For the first time, we demonstrate the feasibility of achieving state-of-the-art performance across diverse applications using a single scalable framework (including 1st place in the VLM3D challenge), eliminating the need for separate task-specific models.
A methodology for clinically driven interactive segmentation evaluation
Oct 10, 2025Abstract:Interactive segmentation is a promising strategy for building robust, generalisable algorithms for volumetric medical image segmentation. However, inconsistent and clinically unrealistic evaluation hinders fair comparison and misrepresents real-world performance. We propose a clinically grounded methodology for defining evaluation tasks and metrics, and built a software framework for constructing standardised evaluation pipelines. We evaluate state-of-the-art algorithms across heterogeneous and complex tasks and observe that (i) minimising information loss when processing user interactions is critical for model robustness, (ii) adaptive-zooming mechanisms boost robustness and speed convergence, (iii) performance drops if validation prompting behaviour/budgets differ from training, (iv) 2D methods perform well with slab-like images and coarse targets, but 3D context helps with large or irregularly shaped targets, (v) performance of non-medical-domain models (e.g. SAM2) degrades with poor contrast and complex shapes.
VIViT: Variable-Input Vision Transformer Framework for 3D MR Image Segmentation
May 13, 2025Abstract:Self-supervised pretrain techniques have been widely used to improve the downstream tasks' performance. However, real-world magnetic resonance (MR) studies usually consist of different sets of contrasts due to different acquisition protocols, which poses challenges for the current deep learning methods on large-scale pretrain and different downstream tasks with different input requirements, since these methods typically require a fixed set of input modalities or, contrasts. To address this challenge, we propose variable-input ViT (VIViT), a transformer-based framework designed for self-supervised pretraining and segmentation finetuning for variable contrasts in each study. With this ability, our approach can maximize the data availability in pretrain, and can transfer the learned knowledge from pretrain to downstream tasks despite variations in input requirements. We validate our method on brain infarct and brain tumor segmentation, where our method outperforms current CNN and ViT-based models with a mean Dice score of 0.624 and 0.883 respectively. These results highlight the efficacy of our design for better adaptability and performance on tasks with real-world heterogeneous MR data.
Multi-Plane Vision Transformer for Hemorrhage Classification Using Axial and Sagittal MRI Data
May 12, 2025Abstract:Identifying brain hemorrhages from magnetic resonance imaging (MRI) is a critical task for healthcare professionals. The diverse nature of MRI acquisitions with varying contrasts and orientation introduce complexity in identifying hemorrhage using neural networks. For acquisitions with varying orientations, traditional methods often involve resampling images to a fixed plane, which can lead to information loss. To address this, we propose a 3D multi-plane vision transformer (MP-ViT) for hemorrhage classification with varying orientation data. It employs two separate transformer encoders for axial and sagittal contrasts, using cross-attention to integrate information across orientations. MP-ViT also includes a modality indication vector to provide missing contrast information to the model. The effectiveness of the proposed model is demonstrated with extensive experiments on real world clinical dataset consists of 10,084 training, 1,289 validation and 1,496 test subjects. MP-ViT achieved substantial improvement in area under the curve (AUC), outperforming the vision transformer (ViT) by 5.5% and CNN-based architectures by 1.8%. These results highlight the potential of MP-ViT in improving performance for hemorrhage detection when different orientation contrasts are needed.
AdaViT: Adaptive Vision Transformer for Flexible Pretrain and Finetune with Variable 3D Medical Image Modalities
Apr 04, 2025



Abstract:Pretrain techniques, whether supervised or self-supervised, are widely used in deep learning to enhance model performance. In real-world clinical scenarios, different sets of magnetic resonance (MR) contrasts are often acquired for different subjects/cases, creating challenges for deep learning models assuming consistent input modalities among all the cases and between pretrain and finetune. Existing methods struggle to maintain performance when there is an input modality/contrast set mismatch with the pretrained model, often resulting in degraded accuracy. We propose an adaptive Vision Transformer (AdaViT) framework capable of handling variable set of input modalities for each case. We utilize a dynamic tokenizer to encode different input image modalities to tokens and take advantage of the characteristics of the transformer to build attention mechanism across variable length of tokens. Through extensive experiments, we demonstrate that this architecture effectively transfers supervised pretrained models to new datasets with different input modality/contrast sets, resulting in superior performance on zero-shot testing, few-shot finetuning, and backward transferring in brain infarct and brain tumor segmentation tasks. Additionally, for self-supervised pretrain, the proposed method is able to maximize the pretrain data and facilitate transferring to diverse downstream tasks with variable sets of input modalities.
A Non-contrast Head CT Foundation Model for Comprehensive Neuro-Trauma Triage
Feb 28, 2025



Abstract:Recent advancements in AI and medical imaging offer transformative potential in emergency head CT interpretation for reducing assessment times and improving accuracy in the face of an increasing request of such scans and a global shortage in radiologists. This study introduces a 3D foundation model for detecting diverse neuro-trauma findings with high accuracy and efficiency. Using large language models (LLMs) for automatic labeling, we generated comprehensive multi-label annotations for critical conditions. Our approach involved pretraining neural networks for hemorrhage subtype segmentation and brain anatomy parcellation, which were integrated into a pretrained comprehensive neuro-trauma detection network through multimodal fine-tuning. Performance evaluation against expert annotations and comparison with CT-CLIP demonstrated strong triage accuracy across major neuro-trauma findings, such as hemorrhage and midline shift, as well as less frequent critical conditions such as cerebral edema and arterial hyperdensity. The integration of neuro-specific features significantly enhanced diagnostic capabilities, achieving an average AUC of 0.861 for 16 neuro-trauma conditions. This work advances foundation models in medical imaging, serving as a benchmark for future AI-assisted neuro-trauma diagnostics in emergency radiology.
Self Pre-training with Adaptive Mask Autoencoders for Variable-Contrast 3D Medical Imaging
Jan 15, 2025Abstract:The Masked Autoencoder (MAE) has recently demonstrated effectiveness in pre-training Vision Transformers (ViT) for analyzing natural images. By reconstructing complete images from partially masked inputs, the ViT encoder gathers contextual information to predict the missing regions. This capability to aggregate context is especially important in medical imaging, where anatomical structures are functionally and mechanically linked to surrounding regions. However, current methods do not consider variations in the number of input images, which is typically the case in real-world Magnetic Resonance (MR) studies. To address this limitation, we propose a 3D Adaptive Masked Autoencoders (AMAE) architecture that accommodates a variable number of 3D input contrasts per subject. A magnetic resonance imaging (MRI) dataset of 45,364 subjects was used for pretraining and a subset of 1648 training, 193 validation and 215 test subjects were used for finetuning. The performance demonstrates that self pre-training of this adaptive masked autoencoders can enhance the infarct segmentation performance by 2.8%-3.7% for ViT-based segmentation models.
Quantifying and Leveraging Predictive Uncertainty for Medical Image Assessment
Jul 08, 2020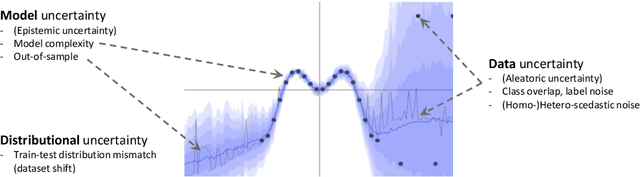

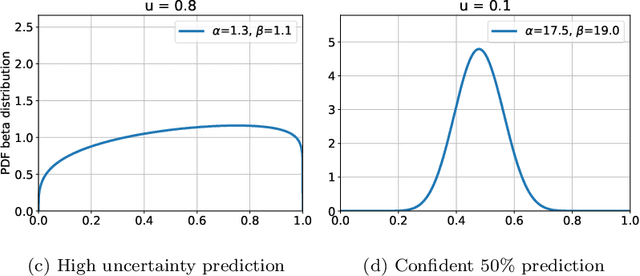
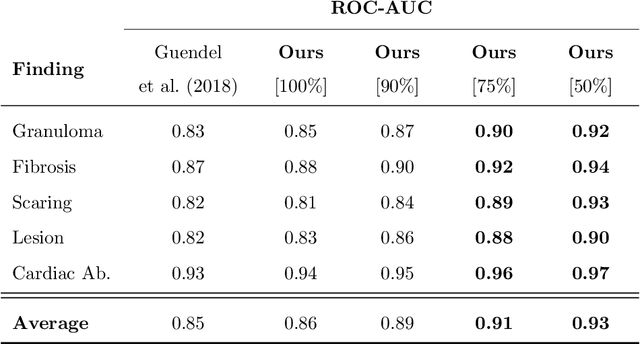
Abstract:The interpretation of medical images is a challenging task, often complicated by the presence of artifacts, occlusions, limited contrast and more. Most notable is the case of chest radiography, where there is a high inter-rater variability in the detection and classification of abnormalities. This is largely due to inconclusive evidence in the data or subjective definitions of disease appearance. An additional example is the classification of anatomical views based on 2D Ultrasound images. Often, the anatomical context captured in a frame is not sufficient to recognize the underlying anatomy. Current machine learning solutions for these problems are typically limited to providing probabilistic predictions, relying on the capacity of underlying models to adapt to limited information and the high degree of label noise. In practice, however, this leads to overconfident systems with poor generalization on unseen data. To account for this, we propose a system that learns not only the probabilistic estimate for classification, but also an explicit uncertainty measure which captures the confidence of the system in the predicted output. We argue that this approach is essential to account for the inherent ambiguity characteristic of medical images from different radiologic exams including computed radiography, ultrasonography and magnetic resonance imaging. In our experiments we demonstrate that sample rejection based on the predicted uncertainty can significantly improve the ROC-AUC for various tasks, e.g., by 8% to 0.91 with an expected rejection rate of under 25% for the classification of different abnormalities in chest radiographs. In addition, we show that using uncertainty-driven bootstrapping to filter the training data, one can achieve a significant increase in robustness and accuracy.
Graph Attention Network based Pruning for Reconstructing 3D Liver Vessel Morphology from Contrasted CT Images
Mar 18, 2020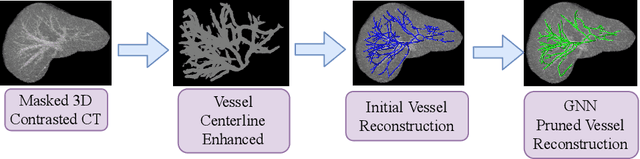
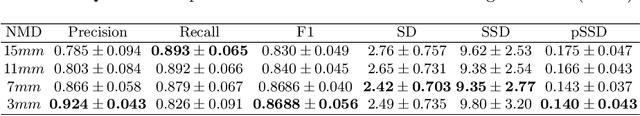
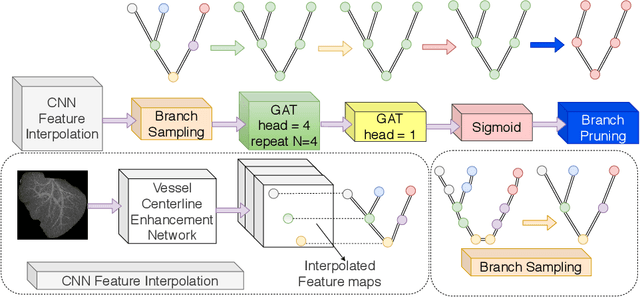
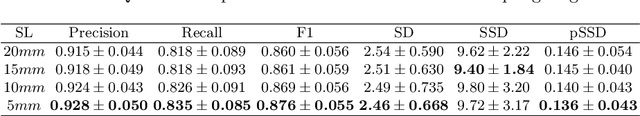
Abstract:With the injection of contrast material into blood vessels, multi-phase contrasted CT images can enhance the visibility of vessel networks in the human body. Reconstructing the 3D geometric morphology of liver vessels from the contrasted CT images can enable multiple liver preoperative surgical planning applications. Automatic reconstruction of liver vessel morphology remains a challenging problem due to the morphological complexity of liver vessels and the inconsistent vessel intensities among different multi-phase contrasted CT images. On the other side, high integrity is required for the 3D reconstruction to avoid decision making biases. In this paper, we propose a framework for liver vessel morphology reconstruction using both a fully convolutional neural network and a graph attention network. A fully convolutional neural network is first trained to produce the liver vessel centerline heatmap. An over-reconstructed liver vessel graph model is then traced based on the heatmap using an image processing based algorithm. We use a graph attention network to prune the false-positive branches by predicting the presence probability of each segmented branch in the initial reconstruction using the aggregated CNN features. We evaluated the proposed framework on an in-house dataset consisting of 418 multi-phase abdomen CT images with contrast. The proposed graph network pruning improves the overall reconstruction F1 score by 6.4% over the baseline. It also outperformed the other state-of-the-art curvilinear structure reconstruction algorithms.
No Surprises: Training Robust Lung Nodule Detection for Low-Dose CT Scans by Augmenting with Adversarial Attacks
Mar 08, 2020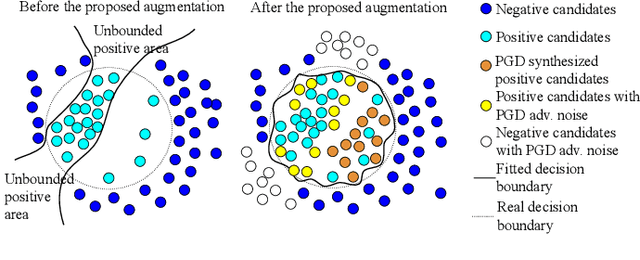
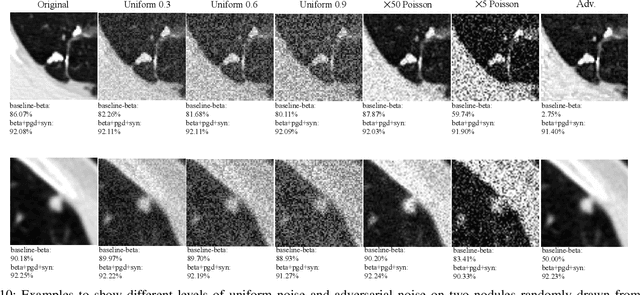
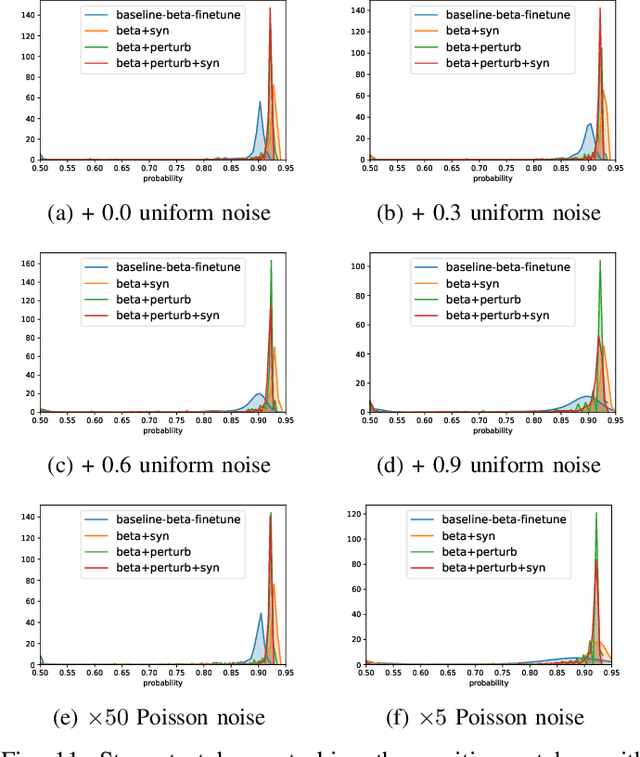
Abstract:Detecting malignant pulmonary nodules at an early stage can allow medical interventions which increases the survival rate of lung cancer patients. Using computer vision techniques to detect nodules can improve the sensitivity and the speed of interpreting chest CT for lung cancer screening. Many studies have used CNNs to detect nodule candidates. Though such approaches have been shown to outperform the conventional image processing based methods regarding the detection accuracy, CNNs are also known to be limited to generalize on under-represented samples in the training set and prone to imperceptible noise perturbations. Such limitations can not be easily addressed by scaling up the dataset or the models. In this work, we propose to add adversarial synthetic nodules and adversarial attack samples to the training data to improve the generalization and the robustness of the lung nodule detection systems. In order to generate hard examples of nodules from a differentiable nodule synthesizer, we use projected gradient descent (PGD) to search the latent code within a bounded neighbourhood that would generate nodules to decrease the detector response. To make the network more robust to unanticipated noise perturbations, we use PGD to search for noise patterns that can trigger the network to give over-confident mistakes. By evaluating on two different benchmark datasets containing consensus annotations from three radiologists, we show that the proposed techniques can improve the detection performance on real CT data. To understand the limitations of both the conventional networks and the proposed augmented networks, we also perform stress-tests on the false positive reduction networks by feeding different types of artificially produced patches. We show that the augmented networks are more robust to both under-represented nodules as well as resistant to noise perturbations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge