Kostas Triaridis
PixCell: A generative foundation model for digital histopathology images
Jun 05, 2025Abstract:The digitization of histology slides has revolutionized pathology, providing massive datasets for cancer diagnosis and research. Contrastive self-supervised and vision-language models have been shown to effectively mine large pathology datasets to learn discriminative representations. On the other hand, generative models, capable of synthesizing realistic and diverse images, present a compelling solution to address unique problems in pathology that involve synthesizing images; overcoming annotated data scarcity, enabling privacy-preserving data sharing, and performing inherently generative tasks, such as virtual staining. We introduce PixCell, the first diffusion-based generative foundation model for histopathology. We train PixCell on PanCan-30M, a vast, diverse dataset derived from 69,184 H\&E-stained whole slide images covering various cancer types. We employ a progressive training strategy and a self-supervision-based conditioning that allows us to scale up training without any annotated data. PixCell generates diverse and high-quality images across multiple cancer types, which we find can be used in place of real data to train a self-supervised discriminative model. Synthetic images shared between institutions are subject to fewer regulatory barriers than would be the case with real clinical images. Furthermore, we showcase the ability to precisely control image generation using a small set of annotated images, which can be used for both data augmentation and educational purposes. Testing on a cell segmentation task, a mask-guided PixCell enables targeted data augmentation, improving downstream performance. Finally, we demonstrate PixCell's ability to use H\&E structural staining to infer results from molecular marker studies; we use this capability to infer IHC staining from H\&E images. Our trained models are publicly released to accelerate research in computational pathology.
Improving Contrastive Learning for Referring Expression Counting
May 28, 2025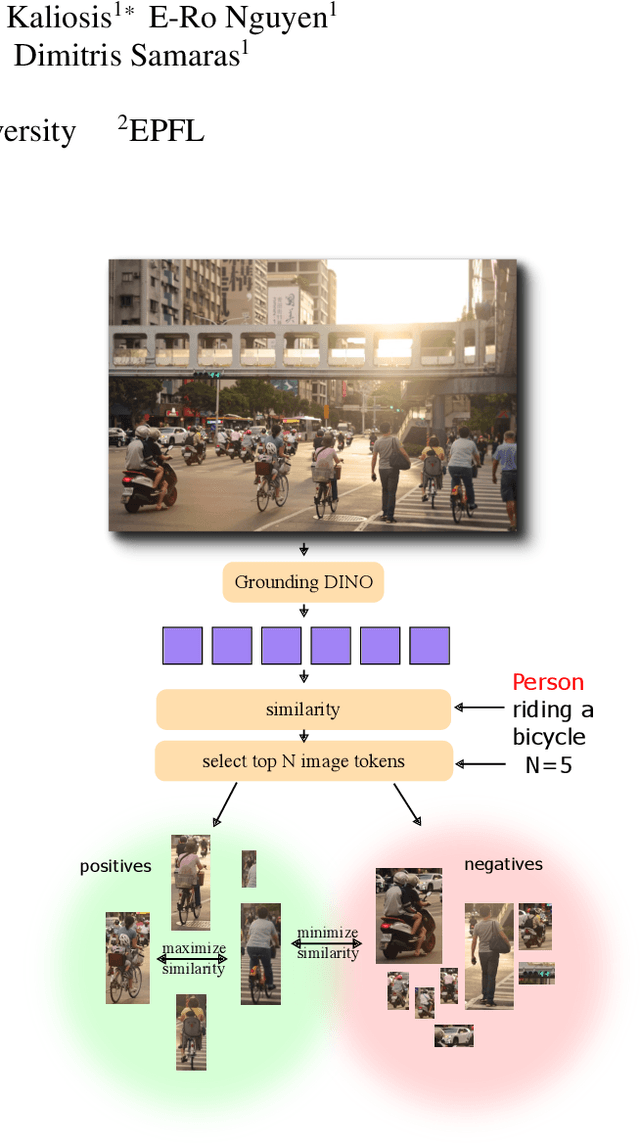
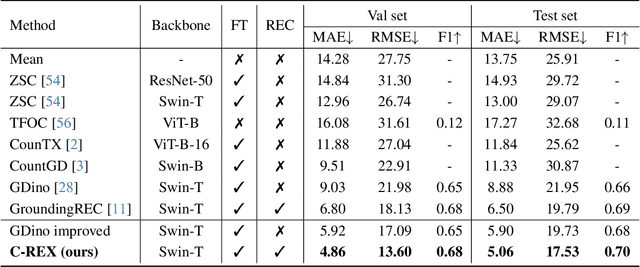

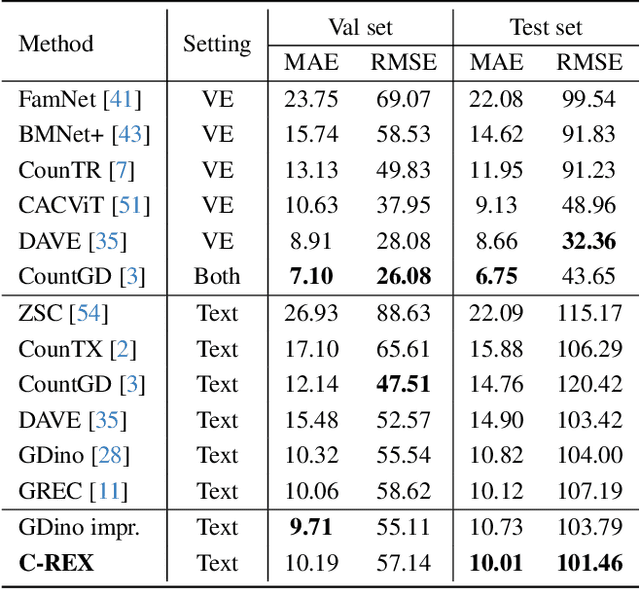
Abstract:Object counting has progressed from class-specific models, which count only known categories, to class-agnostic models that generalize to unseen categories. The next challenge is Referring Expression Counting (REC), where the goal is to count objects based on fine-grained attributes and contextual differences. Existing methods struggle with distinguishing visually similar objects that belong to the same category but correspond to different referring expressions. To address this, we propose C-REX, a novel contrastive learning framework, based on supervised contrastive learning, designed to enhance discriminative representation learning. Unlike prior works, C-REX operates entirely within the image space, avoiding the misalignment issues of image-text contrastive learning, thus providing a more stable contrastive signal. It also guarantees a significantly larger pool of negative samples, leading to improved robustness in the learned representations. Moreover, we showcase that our framework is versatile and generic enough to be applied to other similar tasks like class-agnostic counting. To support our approach, we analyze the key components of sota detection-based models and identify that detecting object centroids instead of bounding boxes is the key common factor behind their success in counting tasks. We use this insight to design a simple yet effective detection-based baseline to build upon. Our experiments show that C-REX achieves state-of-the-art results in REC, outperforming previous methods by more than 22\% in MAE and more than 10\% in RMSE, while also demonstrating strong performance in class-agnostic counting. Code is available at https://github.com/cvlab-stonybrook/c-rex.
Pathology Image Compression with Pre-trained Autoencoders
Mar 14, 2025Abstract:The growing volume of high-resolution Whole Slide Images in digital histopathology poses significant storage, transmission, and computational efficiency challenges. Standard compression methods, such as JPEG, reduce file sizes but often fail to preserve fine-grained phenotypic details critical for downstream tasks. In this work, we repurpose autoencoders (AEs) designed for Latent Diffusion Models as an efficient learned compression framework for pathology images. We systematically benchmark three AE models with varying compression levels and evaluate their reconstruction ability using pathology foundation models. We introduce a fine-tuning strategy to further enhance reconstruction fidelity that optimizes a pathology-specific learned perceptual metric. We validate our approach on downstream tasks, including segmentation, patch classification, and multiple instance learning, showing that replacing images with AE-compressed reconstructions leads to minimal performance degradation. Additionally, we propose a K-means clustering-based quantization method for AE latents, improving storage efficiency while maintaining reconstruction quality. We provide the weights of the fine-tuned autoencoders at https://huggingface.co/collections/StonyBrook-CVLab/pathology-fine-tuned-aes-67d45f223a659ff2e3402dd0.
ZoomLDM: Latent Diffusion Model for multi-scale image generation
Nov 25, 2024Abstract:Diffusion models have revolutionized image generation, yet several challenges restrict their application to large-image domains, such as digital pathology and satellite imagery. Given that it is infeasible to directly train a model on 'whole' images from domains with potential gigapixel sizes, diffusion-based generative methods have focused on synthesizing small, fixed-size patches extracted from these images. However, generating small patches has limited applicability since patch-based models fail to capture the global structures and wider context of large images, which can be crucial for synthesizing (semantically) accurate samples. In this paper, to overcome this limitation, we present ZoomLDM, a diffusion model tailored for generating images across multiple scales. Central to our approach is a novel magnification-aware conditioning mechanism that utilizes self-supervised learning (SSL) embeddings and allows the diffusion model to synthesize images at different 'zoom' levels, i.e., fixed-size patches extracted from large images at varying scales. ZoomLDM achieves state-of-the-art image generation quality across all scales, excelling particularly in the data-scarce setting of generating thumbnails of entire large images. The multi-scale nature of ZoomLDM unlocks additional capabilities in large image generation, enabling computationally tractable and globally coherent image synthesis up to $4096 \times 4096$ pixels and $4\times$ super-resolution. Additionally, multi-scale features extracted from ZoomLDM are highly effective in multiple instance learning experiments. We provide high-resolution examples of the generated images on our website https://histodiffusion.github.io/docs/publications/zoomldm/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge