Debdoot Sheet
SERPENT-VLM : Self-Refining Radiology Report Generation Using Vision Language Models
Apr 27, 2024Abstract:Radiology Report Generation (R2Gen) demonstrates how Multi-modal Large Language Models (MLLMs) can automate the creation of accurate and coherent radiological reports. Existing methods often hallucinate details in text-based reports that don't accurately reflect the image content. To mitigate this, we introduce a novel strategy, SERPENT-VLM (SElf Refining Radiology RePort GENeraTion using Vision Language Models), which improves the R2Gen task by integrating a self-refining mechanism into the MLLM framework. We employ a unique self-supervised loss that leverages similarity between pooled image representations and the contextual representations of the generated radiological text, alongside the standard Causal Language Modeling objective, to refine image-text representations. This allows the model to scrutinize and align the generated text through dynamic interaction between a given image and the generated text, therefore reducing hallucination and continuously enhancing nuanced report generation. SERPENT-VLM outperforms existing baselines such as LLaVA-Med, BiomedGPT, etc., achieving SoTA performance on the IU X-ray and Radiology Objects in COntext (ROCO) datasets, and also proves to be robust against noisy images. A qualitative case study emphasizes the significant advancements towards more sophisticated MLLM frameworks for R2Gen, opening paths for further research into self-supervised refinement in the medical imaging domain.
Knowledge Distillation of Convolutional Neural Networks through Feature Map Transformation using Decision Trees
Mar 10, 2024



Abstract:The interpretation of reasoning by Deep Neural Networks (DNN) is still challenging due to their perceived black-box nature. Therefore, deploying DNNs in several real-world tasks is restricted by the lack of transparency of these models. We propose a distillation approach by extracting features from the final layer of the convolutional neural network (CNN) to address insights to its reasoning. The feature maps in the final layer of a CNN are transformed into a one-dimensional feature vector using a fully connected layer. Subsequently, the extracted features are used to train a decision tree to achieve the best accuracy under constraints of depth and nodes. We use the medical images of dermaMNIST, octMNIST, and pneumoniaMNIST from the medical MNIST datasets to demonstrate our proposed work. We observed that performance of the decision tree is as good as a CNN with minimum complexity. The results encourage interpreting decisions made by the CNNs using decision trees.
CholecTriplet2022: Show me a tool and tell me the triplet -- an endoscopic vision challenge for surgical action triplet detection
Feb 13, 2023



Abstract:Formalizing surgical activities as triplets of the used instruments, actions performed, and target anatomies is becoming a gold standard approach for surgical activity modeling. The benefit is that this formalization helps to obtain a more detailed understanding of tool-tissue interaction which can be used to develop better Artificial Intelligence assistance for image-guided surgery. Earlier efforts and the CholecTriplet challenge introduced in 2021 have put together techniques aimed at recognizing these triplets from surgical footage. Estimating also the spatial locations of the triplets would offer a more precise intraoperative context-aware decision support for computer-assisted intervention. This paper presents the CholecTriplet2022 challenge, which extends surgical action triplet modeling from recognition to detection. It includes weakly-supervised bounding box localization of every visible surgical instrument (or tool), as the key actors, and the modeling of each tool-activity in the form of <instrument, verb, target> triplet. The paper describes a baseline method and 10 new deep learning algorithms presented at the challenge to solve the task. It also provides thorough methodological comparisons of the methods, an in-depth analysis of the obtained results, their significance, and useful insights for future research directions and applications in surgery.
Verifiable and Energy Efficient Medical Image Analysis with Quantised Self-attentive Deep Neural Networks
Sep 30, 2022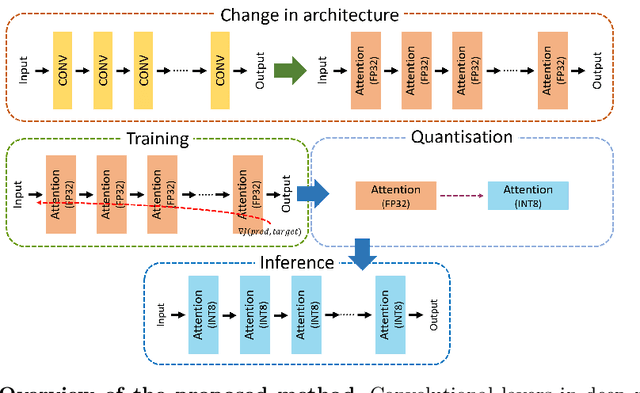
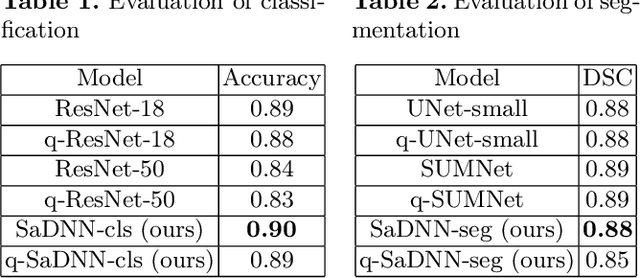
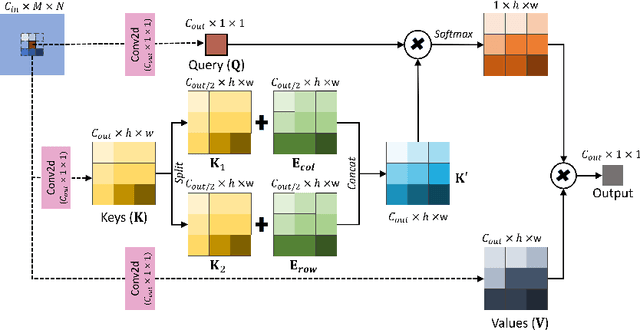
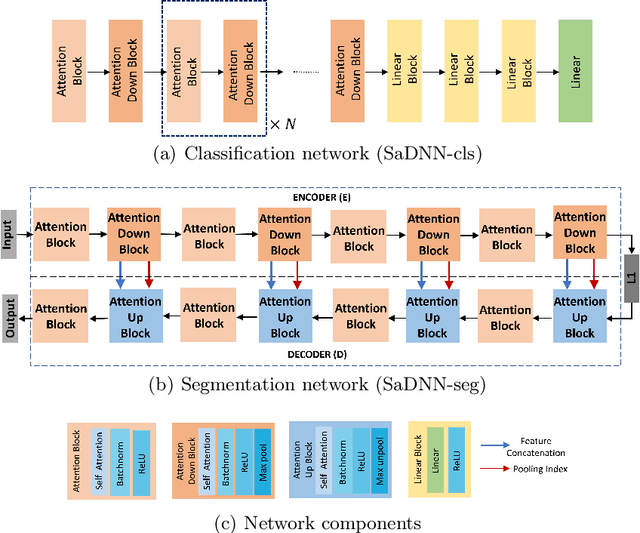
Abstract:Convolutional Neural Networks have played a significant role in various medical imaging tasks like classification and segmentation. They provide state-of-the-art performance compared to classical image processing algorithms. However, the major downside of these methods is the high computational complexity, reliance on high-performance hardware like GPUs and the inherent black-box nature of the model. In this paper, we propose quantised stand-alone self-attention based models as an alternative to traditional CNNs. In the proposed class of networks, convolutional layers are replaced with stand-alone self-attention layers, and the network parameters are quantised after training. We experimentally validate the performance of our method on classification and segmentation tasks. We observe a $50-80\%$ reduction in model size, $60-80\%$ lesser number of parameters, $40-85\%$ fewer FLOPs and $65-80\%$ more energy efficiency during inference on CPUs. The code will be available at \href {https://github.com/Rakshith2597/Quantised-Self-Attentive-Deep-Neural-Network}{https://github.com/Rakshith2597/Quantised-Self-Attentive-Deep-Neural-Network}.
CholecTriplet2021: A benchmark challenge for surgical action triplet recognition
Apr 10, 2022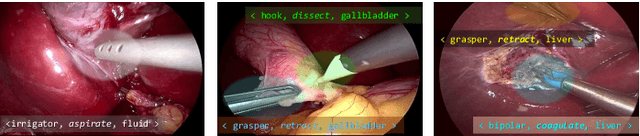
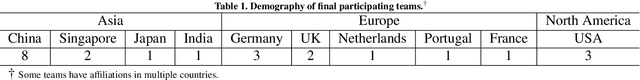
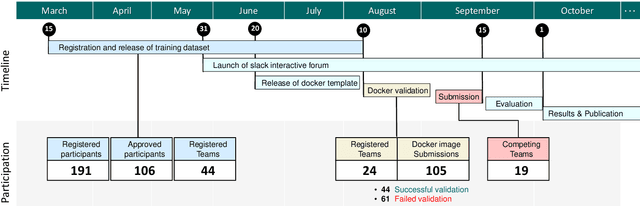
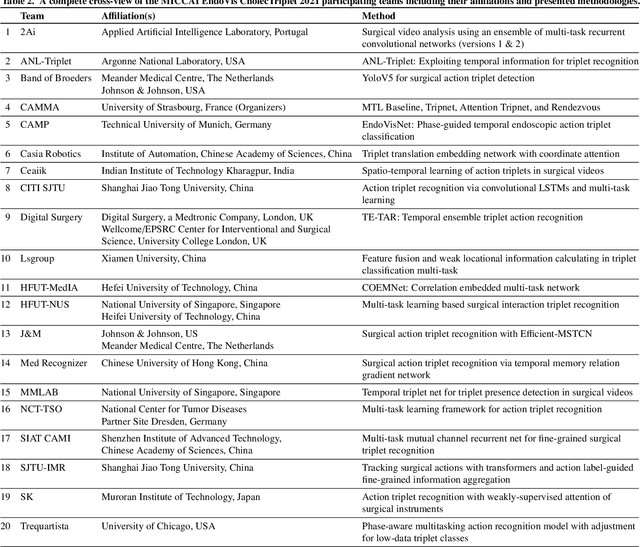
Abstract:Context-aware decision support in the operating room can foster surgical safety and efficiency by leveraging real-time feedback from surgical workflow analysis. Most existing works recognize surgical activities at a coarse-grained level, such as phases, steps or events, leaving out fine-grained interaction details about the surgical activity; yet those are needed for more helpful AI assistance in the operating room. Recognizing surgical actions as triplets of <instrument, verb, target> combination delivers comprehensive details about the activities taking place in surgical videos. This paper presents CholecTriplet2021: an endoscopic vision challenge organized at MICCAI 2021 for the recognition of surgical action triplets in laparoscopic videos. The challenge granted private access to the large-scale CholecT50 dataset, which is annotated with action triplet information. In this paper, we present the challenge setup and assessment of the state-of-the-art deep learning methods proposed by the participants during the challenge. A total of 4 baseline methods from the challenge organizers and 19 new deep learning algorithms by competing teams are presented to recognize surgical action triplets directly from surgical videos, achieving mean average precision (mAP) ranging from 4.2% to 38.1%. This study also analyzes the significance of the results obtained by the presented approaches, performs a thorough methodological comparison between them, in-depth result analysis, and proposes a novel ensemble method for enhanced recognition. Our analysis shows that surgical workflow analysis is not yet solved, and also highlights interesting directions for future research on fine-grained surgical activity recognition which is of utmost importance for the development of AI in surgery.
Lung Segmentation and Nodule Detection in Computed Tomography Scan using a Convolutional Neural Network Trained Adversarially using Turing Test Loss
Jun 16, 2020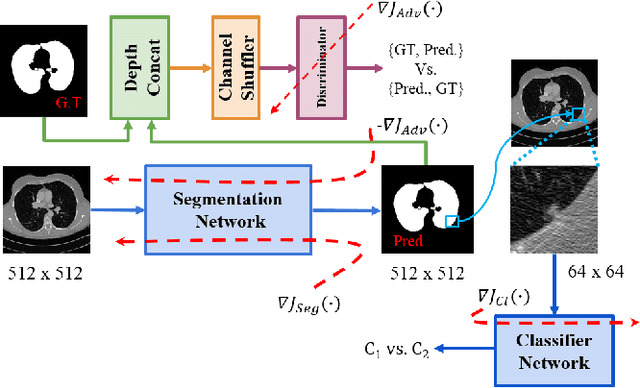
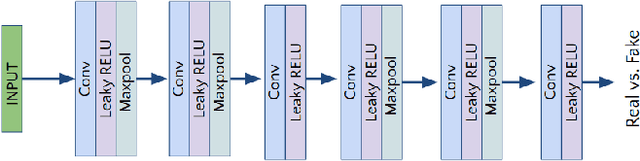
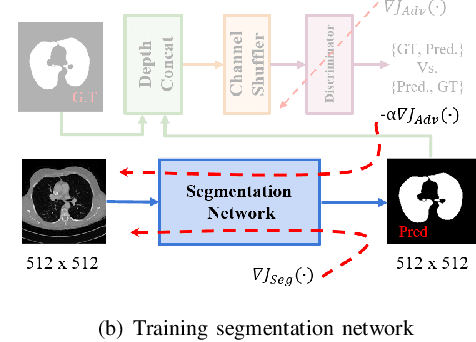
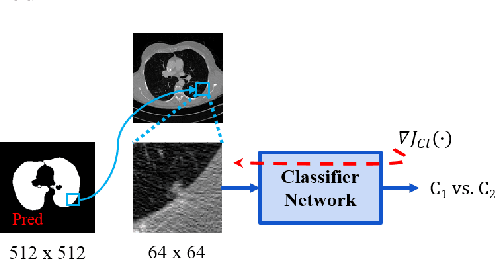
Abstract:Lung cancer is the most common form of cancer found worldwide with a high mortality rate. Early detection of pulmonary nodules by screening with a low-dose computed tomography (CT) scan is crucial for its effective clinical management. Nodules which are symptomatic of malignancy occupy about 0.0125 - 0.025\% of volume in a CT scan of a patient. Manual screening of all slices is a tedious task and presents a high risk of human errors. To tackle this problem we propose a computationally efficient two stage framework. In the first stage, a convolutional neural network (CNN) trained adversarially using Turing test loss segments the lung region. In the second stage, patches sampled from the segmented region are then classified to detect the presence of nodules. The proposed method is experimentally validated on the LUNA16 challenge dataset with a dice coefficient of $0.984\pm0.0007$ for 10-fold cross-validation.
Identification of Cervical Pathology using Adversarial Neural Networks
Apr 28, 2020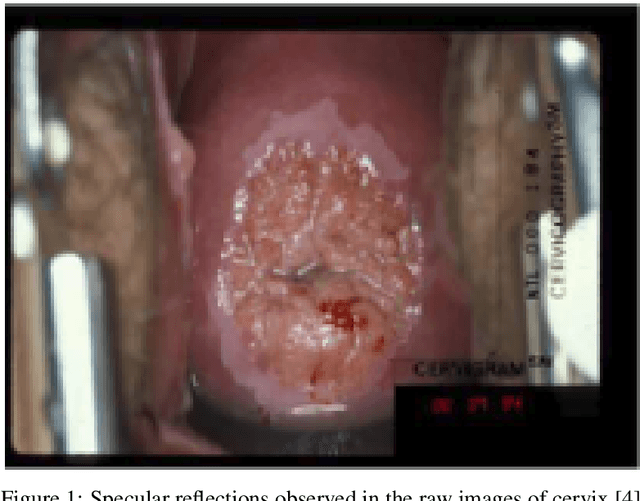
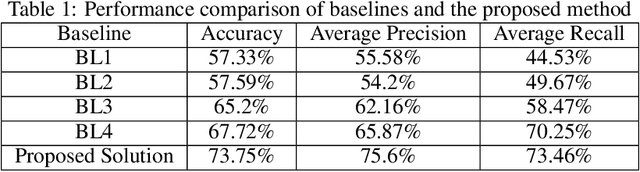
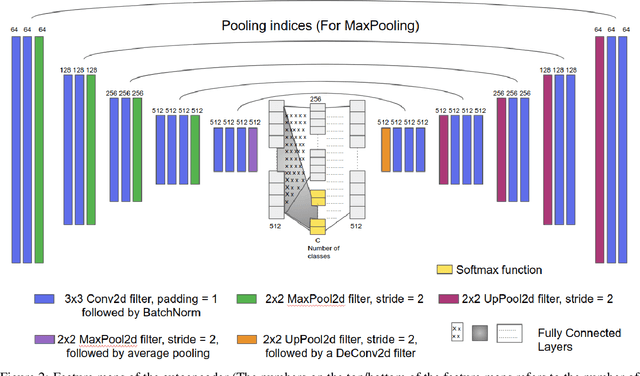
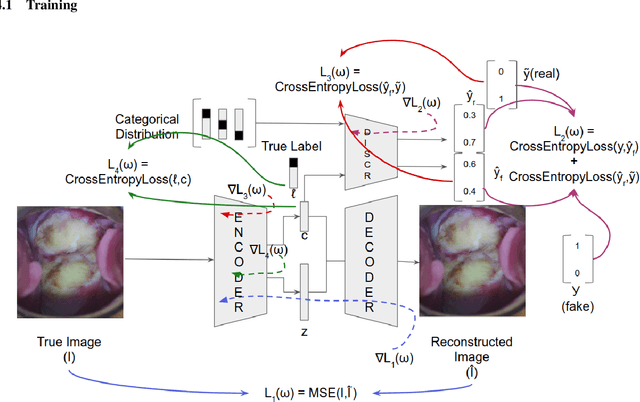
Abstract:Various screening and diagnostic methods have led to a large reduction of cervical cancer death rates in developed countries. However, cervical cancer is the leading cause of cancer related deaths in women in India and other low and middle income countries (LMICs) especially among the urban poor and slum dwellers. Several sophisticated techniques such as cytology tests, HPV tests etc. have been widely used for screening of cervical cancer. These tests are inherently time consuming. In this paper, we propose a convolutional autoencoder based framework, having an architecture similar to SegNet which is trained in an adversarial fashion for classifying images of the cervix acquired using a colposcope. We validate performance on the Intel-Mobile ODT cervical image classification dataset. The proposed method outperforms the standard technique of fine-tuning convolutional neural networks pre-trained on ImageNet database with an average accuracy of 73.75%.
IROS 2019 Lifelong Robotic Vision Challenge -- Lifelong Object Recognition Report
Apr 26, 2020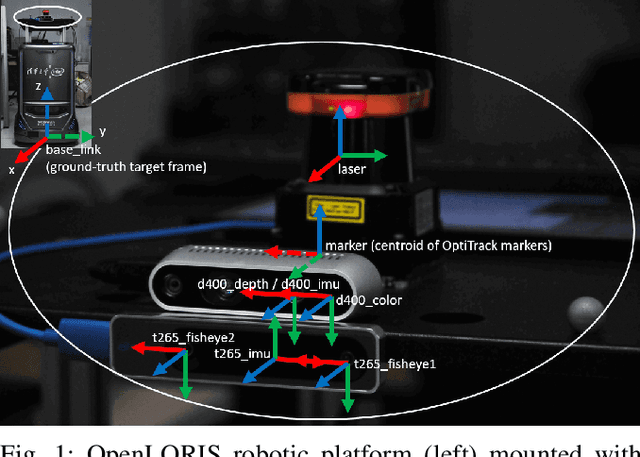
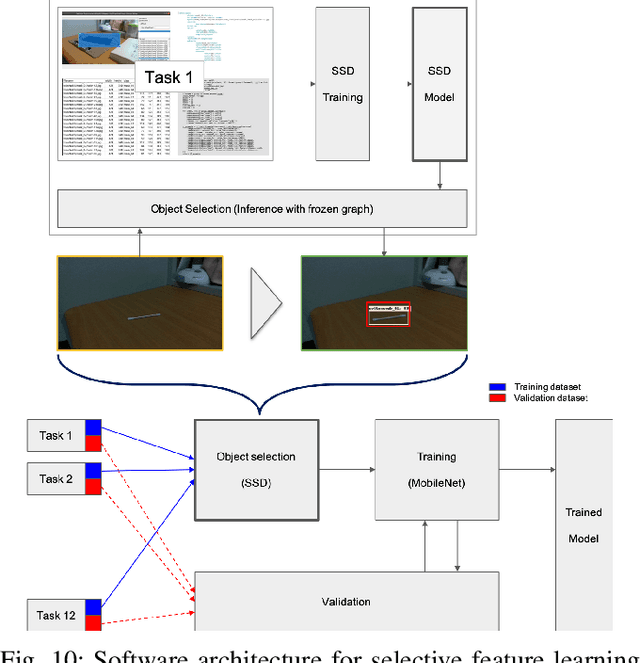

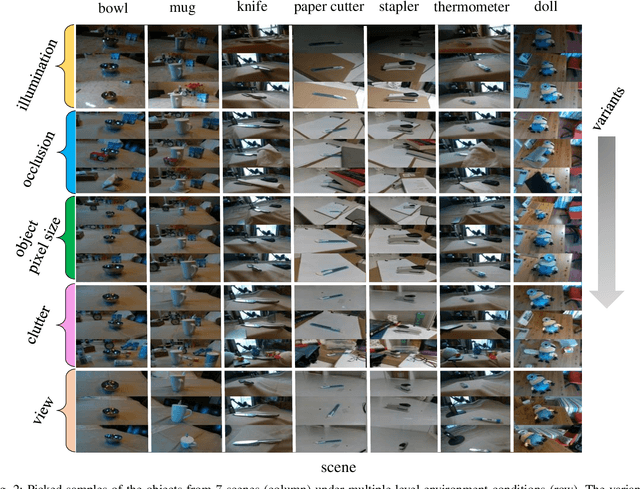
Abstract:This report summarizes IROS 2019-Lifelong Robotic Vision Competition (Lifelong Object Recognition Challenge) with methods and results from the top $8$ finalists (out of over~$150$ teams). The competition dataset (L)ifel(O)ng (R)obotic V(IS)ion (OpenLORIS) - Object Recognition (OpenLORIS-object) is designed for driving lifelong/continual learning research and application in robotic vision domain, with everyday objects in home, office, campus, and mall scenarios. The dataset explicitly quantifies the variants of illumination, object occlusion, object size, camera-object distance/angles, and clutter information. Rules are designed to quantify the learning capability of the robotic vision system when faced with the objects appearing in the dynamic environments in the contest. Individual reports, dataset information, rules, and released source code can be found at the project homepage: "https://lifelong-robotic-vision.github.io/competition/".
A Two-Stage Multiple Instance Learning Framework for the Detection of Breast Cancer in Mammograms
Apr 24, 2020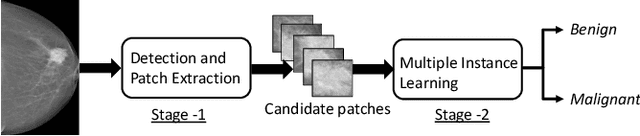
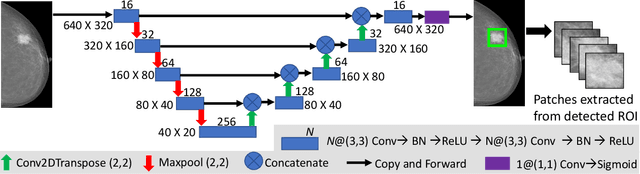
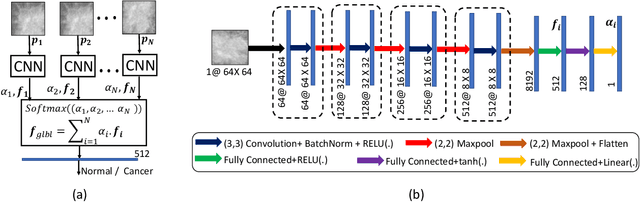
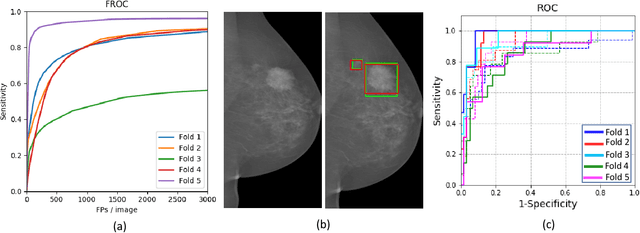
Abstract:Mammograms are commonly employed in the large scale screening of breast cancer which is primarily characterized by the presence of malignant masses. However, automated image-level detection of malignancy is a challenging task given the small size of the mass regions and difficulty in discriminating between malignant, benign mass and healthy dense fibro-glandular tissue. To address these issues, we explore a two-stage Multiple Instance Learning (MIL) framework. A Convolutional Neural Network (CNN) is trained in the first stage to extract local candidate patches in the mammograms that may contain either a benign or malignant mass. The second stage employs a MIL strategy for an image level benign vs. malignant classification. A global image-level feature is computed as a weighted average of patch-level features learned using a CNN. Our method performed well on the task of localization of masses with an average Precision/Recall of 0.76/0.80 and acheived an average AUC of 0.91 on the imagelevel classification task using a five-fold cross-validation on the INbreast dataset. Restricting the MIL only to the candidate patches extracted in Stage 1 led to a significant improvement in classification performance in comparison to a dense extraction of patches from the entire mammogram.
Learning Decision Ensemble using a Graph Neural Network for Comorbidity Aware Chest Radiograph Screening
Apr 24, 2020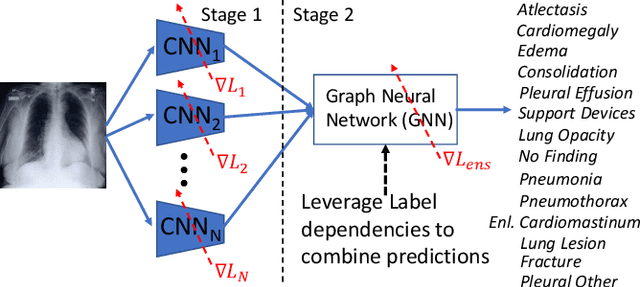
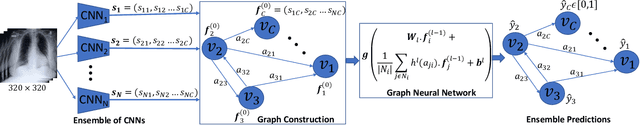
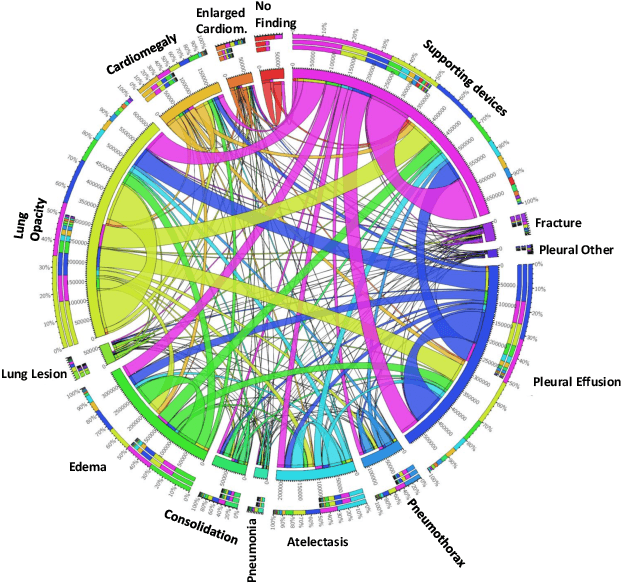
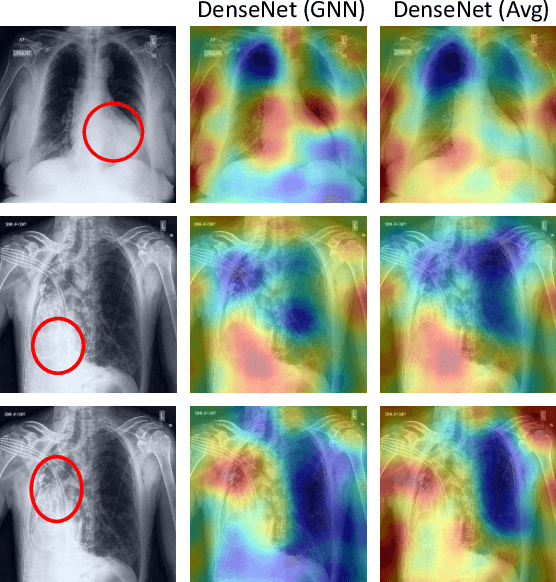
Abstract:Chest radiographs are primarily employed for the screening of cardio, thoracic and pulmonary conditions. Machine learning based automated solutions are being developed to reduce the burden of routine screening on Radiologists, allowing them to focus on critical cases. While recent efforts demonstrate the use of ensemble of deep convolutional neural networks(CNN), they do not take disease comorbidity into consideration, thus lowering their screening performance. To address this issue, we propose a Graph Neural Network (GNN) based solution to obtain ensemble predictions which models the dependencies between different diseases. A comprehensive evaluation of the proposed method demonstrated its potential by improving the performance over standard ensembling technique across a wide range of ensemble constructions. The best performance was achieved using the GNN ensemble of DenseNet121 with an average AUC of 0.821 across thirteen disease comorbidities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge