Arunava Chakravarty
Stochastic Siamese MAE Pretraining for Longitudinal Medical Images
Dec 29, 2025Abstract:Temporally aware image representations are crucial for capturing disease progression in 3D volumes of longitudinal medical datasets. However, recent state-of-the-art self-supervised learning approaches like Masked Autoencoding (MAE), despite their strong representation learning capabilities, lack temporal awareness. In this paper, we propose STAMP (Stochastic Temporal Autoencoder with Masked Pretraining), a Siamese MAE framework that encodes temporal information through a stochastic process by conditioning on the time difference between the 2 input volumes. Unlike deterministic Siamese approaches, which compare scans from different time points but fail to account for the inherent uncertainty in disease evolution, STAMP learns temporal dynamics stochastically by reframing the MAE reconstruction loss as a conditional variational inference objective. We evaluated STAMP on two OCT and one MRI datasets with multiple visits per patient. STAMP pretrained ViT models outperformed both existing temporal MAE methods and foundation models on different late stage Age-Related Macular Degeneration and Alzheimer's Disease progression prediction which require models to learn the underlying non-deterministic temporal dynamics of the diseases.
Forecasting Disease Progression with Parallel Hyperplanes in Longitudinal Retinal OCT
Sep 30, 2024



Abstract:Predicting future disease progression risk from medical images is challenging due to patient heterogeneity, and subtle or unknown imaging biomarkers. Moreover, deep learning (DL) methods for survival analysis are susceptible to image domain shifts across scanners. We tackle these issues in the task of predicting late dry Age-related Macular Degeneration (dAMD) onset from retinal OCT scans. We propose a novel DL method for survival prediction to jointly predict from the current scan a risk score, inversely related to time-to-conversion, and the probability of conversion within a time interval $t$. It uses a family of parallel hyperplanes generated by parameterizing the bias term as a function of $t$. In addition, we develop unsupervised losses based on intra-subject image pairs to ensure that risk scores increase over time and that future conversion predictions are consistent with AMD stage prediction using actual scans of future visits. Such losses enable data-efficient fine-tuning of the trained model on new unlabeled datasets acquired with a different scanner. Extensive evaluation on two large datasets acquired with different scanners resulted in a mean AUROCs of 0.82 for Dataset-1 and 0.83 for Dataset-2, across prediction intervals of 6,12 and 24 months.
Time-Equivariant Contrastive Learning for Degenerative Disease Progression in Retinal OCT
May 15, 2024



Abstract:Contrastive pretraining provides robust representations by ensuring their invariance to different image transformations while simultaneously preventing representational collapse. Equivariant contrastive learning, on the other hand, provides representations sensitive to specific image transformations while remaining invariant to others. By introducing equivariance to time-induced transformations, such as disease-related anatomical changes in longitudinal imaging, the model can effectively capture such changes in the representation space. In this work, we pro-pose a Time-equivariant Contrastive Learning (TC) method. First, an encoder embeds two unlabeled scans from different time points of the same patient into the representation space. Next, a temporal equivariance module is trained to predict the representation of a later visit based on the representation from one of the previous visits and the corresponding time interval with a novel regularization loss term while preserving the invariance property to irrelevant image transformations. On a large longitudinal dataset, our model clearly outperforms existing equivariant contrastive methods in predicting progression from intermediate age-related macular degeneration (AMD) to advanced wet-AMD within a specified time-window.
3DTINC: Time-Equivariant Non-Contrastive Learning for Predicting Disease Progression from Longitudinal OCTs
Dec 28, 2023



Abstract:Self-supervised learning (SSL) has emerged as a powerful technique for improving the efficiency and effectiveness of deep learning models. Contrastive methods are a prominent family of SSL that extract similar representations of two augmented views of an image while pushing away others in the representation space as negatives. However, the state-of-the-art contrastive methods require large batch sizes and augmentations designed for natural images that are impractical for 3D medical images. To address these limitations, we propose a new longitudinal SSL method, 3DTINC, based on non-contrastive learning. It is designed to learn perturbation-invariant features for 3D optical coherence tomography (OCT) volumes, using augmentations specifically designed for OCT. We introduce a new non-contrastive similarity loss term that learns temporal information implicitly from intra-patient scans acquired at different times. Our experiments show that this temporal information is crucial for predicting progression of retinal diseases, such as age-related macular degeneration (AMD). After pretraining with 3DTINC, we evaluated the learned representations and the prognostic models on two large-scale longitudinal datasets of retinal OCTs where we predict the conversion to wet-AMD within a six months interval. Our results demonstrate that each component of our contributions is crucial for learning meaningful representations useful in predicting disease progression from longitudinal volumetric scans.
Pretrained Deep 2.5D Models for Efficient Predictive Modeling from Retinal OCT
Jul 25, 2023Abstract:In the field of medical imaging, 3D deep learning models play a crucial role in building powerful predictive models of disease progression. However, the size of these models presents significant challenges, both in terms of computational resources and data requirements. Moreover, achieving high-quality pretraining of 3D models proves to be even more challenging. To address these issues, hybrid 2.5D approaches provide an effective solution for utilizing 3D volumetric data efficiently using 2D models. Combining 2D and 3D techniques offers a promising avenue for optimizing performance while minimizing memory requirements. In this paper, we explore 2.5D architectures based on a combination of convolutional neural networks (CNNs), long short-term memory (LSTM), and Transformers. In addition, leveraging the benefits of recent non-contrastive pretraining approaches in 2D, we enhanced the performance and data efficiency of 2.5D techniques even further. We demonstrate the effectiveness of architectures and associated pretraining on a task of predicting progression to wet age-related macular degeneration (AMD) within a six-month period on two large longitudinal OCT datasets.
Morph-SSL: Self-Supervision with Longitudinal Morphing to Predict AMD Progression from OCT
Apr 17, 2023



Abstract:The lack of reliable biomarkers makes predicting the conversion from intermediate to neovascular age-related macular degeneration (iAMD, nAMD) a challenging task. We develop a Deep Learning (DL) model to predict the future risk of conversion of an eye from iAMD to nAMD from its current OCT scan. Although eye clinics generate vast amounts of longitudinal OCT scans to monitor AMD progression, only a small subset can be manually labeled for supervised DL. To address this issue, we propose Morph-SSL, a novel Self-supervised Learning (SSL) method for longitudinal data. It uses pairs of unlabelled OCT scans from different visits and involves morphing the scan from the previous visit to the next. The Decoder predicts the transformation for morphing and ensures a smooth feature manifold that can generate intermediate scans between visits through linear interpolation. Next, the Morph-SSL trained features are input to a Classifier which is trained in a supervised manner to model the cumulative probability distribution of the time to conversion with a sigmoidal function. Morph-SSL was trained on unlabelled scans of 399 eyes (3570 visits). The Classifier was evaluated with a five-fold cross-validation on 2418 scans from 343 eyes with clinical labels of the conversion date. The Morph-SSL features achieved an AUC of 0.766 in predicting the conversion to nAMD within the next 6 months, outperforming the same network when trained end-to-end from scratch or pre-trained with popular SSL methods. Automated prediction of the future risk of nAMD onset can enable timely treatment and individualized AMD management.
Learning Spatio-Temporal Model of Disease Progression with NeuralODEs from Longitudinal Volumetric Data
Nov 08, 2022



Abstract:Robust forecasting of the future anatomical changes inflicted by an ongoing disease is an extremely challenging task that is out of grasp even for experienced healthcare professionals. Such a capability, however, is of great importance since it can improve patient management by providing information on the speed of disease progression already at the admission stage, or it can enrich the clinical trials with fast progressors and avoid the need for control arms by the means of digital twins. In this work, we develop a deep learning method that models the evolution of age-related disease by processing a single medical scan and providing a segmentation of the target anatomy at a requested future point in time. Our method represents a time-invariant physical process and solves a large-scale problem of modeling temporal pixel-level changes utilizing NeuralODEs. In addition, we demonstrate the approaches to incorporate the prior domain-specific constraints into our method and define temporal Dice loss for learning temporal objectives. To evaluate the applicability of our approach across different age-related diseases and imaging modalities, we developed and tested the proposed method on the datasets with 967 retinal OCT volumes of 100 patients with Geographic Atrophy, and 2823 brain MRI volumes of 633 patients with Alzheimer's Disease. For Geographic Atrophy, the proposed method outperformed the related baseline models in the atrophy growth prediction. For Alzheimer's Disease, the proposed method demonstrated remarkable performance in predicting the brain ventricle changes induced by the disease, achieving the state-of-the-art result on TADPOLE challenge.
TINC: Temporally Informed Non-Contrastive Learning for Disease Progression Modeling in Retinal OCT Volumes
Jun 30, 2022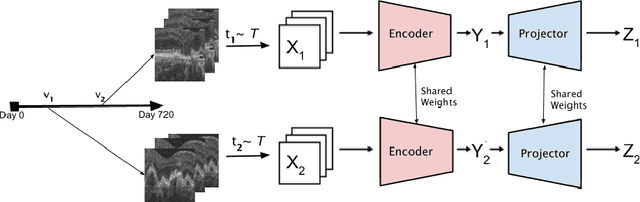
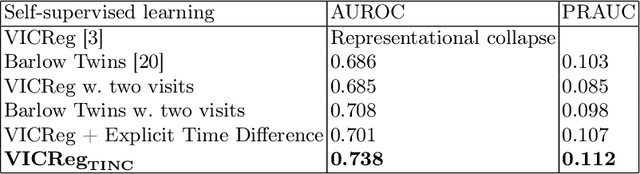

Abstract:Recent contrastive learning methods achieved state-of-the-art in low label regimes. However, the training requires large batch sizes and heavy augmentations to create multiple views of an image. With non-contrastive methods, the negatives are implicitly incorporated in the loss, allowing different images and modalities as pairs. Although the meta-information (i.e., age, sex) in medical imaging is abundant, the annotations are noisy and prone to class imbalance. In this work, we exploited already existing temporal information (different visits from a patient) in a longitudinal optical coherence tomography (OCT) dataset using temporally informed non-contrastive loss (TINC) without increasing complexity and need for negative pairs. Moreover, our novel pair-forming scheme can avoid heavy augmentations and implicitly incorporates the temporal information in the pairs. Finally, these representations learned from the pretraining are more successful in predicting disease progression where the temporal information is crucial for the downstream task. More specifically, our model outperforms existing models in predicting the risk of conversion within a time frame from intermediate age-related macular degeneration (AMD) to the late wet-AMD stage.
A Two-Stage Multiple Instance Learning Framework for the Detection of Breast Cancer in Mammograms
Apr 24, 2020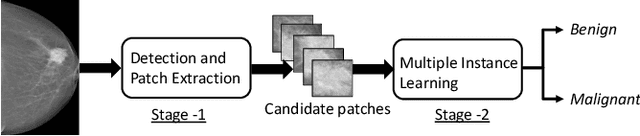
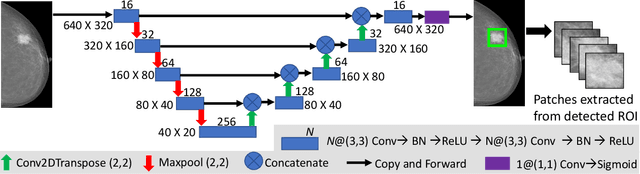
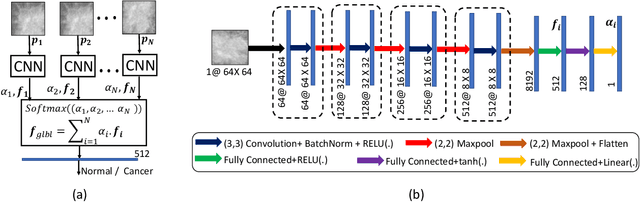
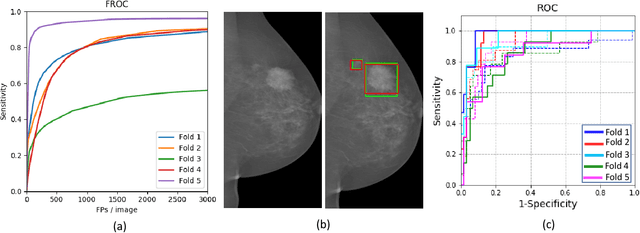
Abstract:Mammograms are commonly employed in the large scale screening of breast cancer which is primarily characterized by the presence of malignant masses. However, automated image-level detection of malignancy is a challenging task given the small size of the mass regions and difficulty in discriminating between malignant, benign mass and healthy dense fibro-glandular tissue. To address these issues, we explore a two-stage Multiple Instance Learning (MIL) framework. A Convolutional Neural Network (CNN) is trained in the first stage to extract local candidate patches in the mammograms that may contain either a benign or malignant mass. The second stage employs a MIL strategy for an image level benign vs. malignant classification. A global image-level feature is computed as a weighted average of patch-level features learned using a CNN. Our method performed well on the task of localization of masses with an average Precision/Recall of 0.76/0.80 and acheived an average AUC of 0.91 on the imagelevel classification task using a five-fold cross-validation on the INbreast dataset. Restricting the MIL only to the candidate patches extracted in Stage 1 led to a significant improvement in classification performance in comparison to a dense extraction of patches from the entire mammogram.
Learning Decision Ensemble using a Graph Neural Network for Comorbidity Aware Chest Radiograph Screening
Apr 24, 2020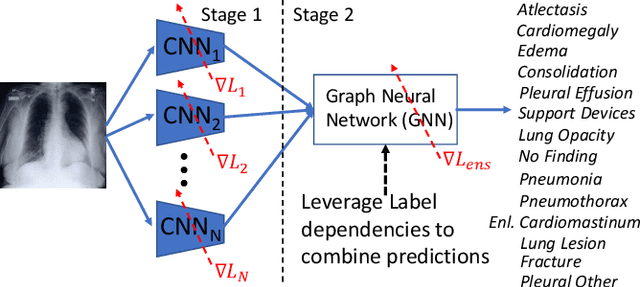
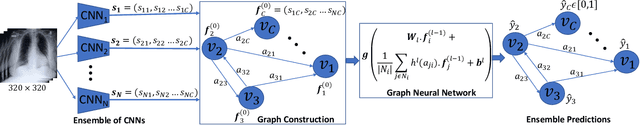
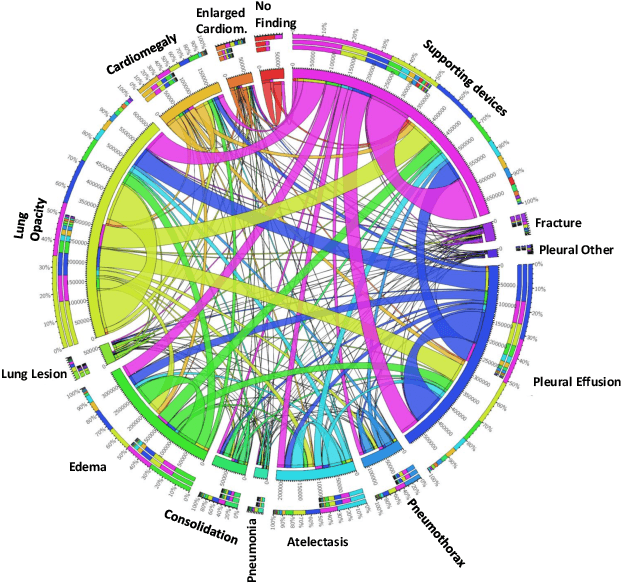
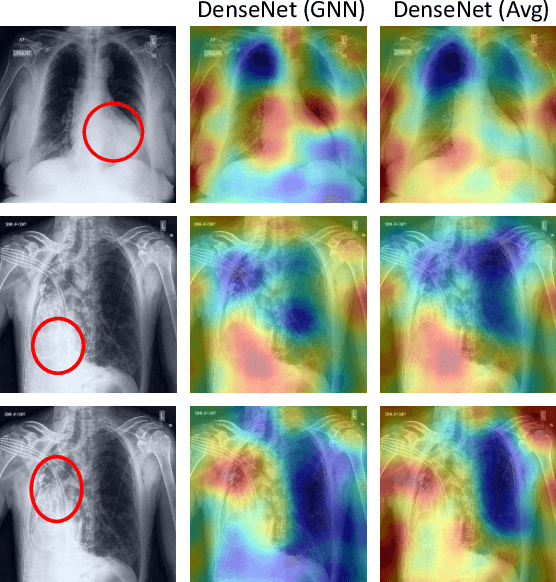
Abstract:Chest radiographs are primarily employed for the screening of cardio, thoracic and pulmonary conditions. Machine learning based automated solutions are being developed to reduce the burden of routine screening on Radiologists, allowing them to focus on critical cases. While recent efforts demonstrate the use of ensemble of deep convolutional neural networks(CNN), they do not take disease comorbidity into consideration, thus lowering their screening performance. To address this issue, we propose a Graph Neural Network (GNN) based solution to obtain ensemble predictions which models the dependencies between different diseases. A comprehensive evaluation of the proposed method demonstrated its potential by improving the performance over standard ensembling technique across a wide range of ensemble constructions. The best performance was achieved using the GNN ensemble of DenseNet121 with an average AUC of 0.821 across thirteen disease comorbidities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge