Zhiwei Wang
CADET: Context-Conditioned Ads CTR Prediction With a Decoder-Only Transformer
Feb 11, 2026Abstract:Click-through rate (CTR) prediction is fundamental to online advertising systems. While Deep Learning Recommendation Models (DLRMs) with explicit feature interactions have long dominated this domain, recent advances in generative recommenders have shown promising results in content recommendation. However, adapting these transformer-based architectures to ads CTR prediction still presents unique challenges, including handling post-scoring contextual signals, maintaining offline-online consistency, and scaling to industrial workloads. We present CADET (Context-Conditioned Ads Decoder-Only Transformer), an end-to-end decoder-only transformer for ads CTR prediction deployed at LinkedIn. Our approach introduces several key innovations: (1) a context-conditioned decoding architecture with multi-tower prediction heads that explicitly model post-scoring signals such as ad position, resolving the chicken-and-egg problem between predicted CTR and ranking; (2) a self-gated attention mechanism that stabilizes training by adaptively regulating information flow at both representation and interaction levels; (3) a timestamp-based variant of Rotary Position Embedding (RoPE) that captures temporal relationships across timescales from seconds to months; (4) session masking strategies that prevent the model from learning dependencies on unavailable in-session events, addressing train-serve skew; and (5) production engineering techniques including tensor packing, sequence chunking, and custom Flash Attention kernels that enable efficient training and serving at scale. In online A/B testing, CADET achieves a 11.04\% CTR lift compared to the production LiRank baseline model, a hybrid ensemble of DCNv2 and sequential encoders. The system has been successfully deployed on LinkedIn's advertising platform, serving the main traffic for homefeed sponsored updates.
Pairing-free Group-level Knowledge Distillation for Robust Gastrointestinal Lesion Classification in White-Light Endoscopy
Jan 14, 2026Abstract:White-Light Imaging (WLI) is the standard for endoscopic cancer screening, but Narrow-Band Imaging (NBI) offers superior diagnostic details. A key challenge is transferring knowledge from NBI to enhance WLI-only models, yet existing methods are critically hampered by their reliance on paired NBI-WLI images of the same lesion, a costly and often impractical requirement that leaves vast amounts of clinical data untapped. In this paper, we break this paradigm by introducing PaGKD, a novel Pairing-free Group-level Knowledge Distillation framework that that enables effective cross-modal learning using unpaired WLI and NBI data. Instead of forcing alignment between individual, often semantically mismatched image instances, PaGKD operates at the group level to distill more complete and compatible knowledge across modalities. Central to PaGKD are two complementary modules: (1) Group-level Prototype Distillation (GKD-Pro) distills compact group representations by extracting modality-invariant semantic prototypes via shared lesion-aware queries; (2) Group-level Dense Distillation (GKD-Den) performs dense cross-modal alignment by guiding group-aware attention with activation-derived relation maps. Together, these modules enforce global semantic consistency and local structural coherence without requiring image-level correspondence. Extensive experiments on four clinical datasets demonstrate that PaGKD consistently and significantly outperforms state-of-the-art methods, achieving relative AUC improvements of 3.3%, 1.1%, 2.8%, and 3.2%, respectively, establishing a new direction for cross-modal learning from unpaired data.
FIA-Edit: Frequency-Interactive Attention for Efficient and High-Fidelity Inversion-Free Text-Guided Image Editing
Nov 15, 2025Abstract:Text-guided image editing has advanced rapidly with the rise of diffusion models. While flow-based inversion-free methods offer high efficiency by avoiding latent inversion, they often fail to effectively integrate source information, leading to poor background preservation, spatial inconsistencies, and over-editing due to the lack of effective integration of source information. In this paper, we present FIA-Edit, a novel inversion-free framework that achieves high-fidelity and semantically precise edits through a Frequency-Interactive Attention. Specifically, we design two key components: (1) a Frequency Representation Interaction (FRI) module that enhances cross-domain alignment by exchanging frequency components between source and target features within self-attention, and (2) a Feature Injection (FIJ) module that explicitly incorporates source-side queries, keys, values, and text embeddings into the target branch's cross-attention to preserve structure and semantics. Comprehensive and extensive experiments demonstrate that FIA-Edit supports high-fidelity editing at low computational cost (~6s per 512 * 512 image on an RTX 4090) and consistently outperforms existing methods across diverse tasks in visual quality, background fidelity, and controllability. Furthermore, we are the first to extend text-guided image editing to clinical applications. By synthesizing anatomically coherent hemorrhage variations in surgical images, FIA-Edit opens new opportunities for medical data augmentation and delivers significant gains in downstream bleeding classification. Our project is available at: https://github.com/kk42yy/FIA-Edit.
MAUGIF: Mechanism-Aware Unsupervised General Image Fusion via Dual Cross-Image Autoencoders
Nov 13, 2025Abstract:Image fusion aims to integrate structural and complementary information from multi-source images. However, existing fusion methods are often either highly task-specific, or general frameworks that apply uniform strategies across diverse tasks, ignoring their distinct fusion mechanisms. To address this issue, we propose a mechanism-aware unsupervised general image fusion (MAUGIF) method based on dual cross-image autoencoders. Initially, we introduce a classification of additive and multiplicative fusion according to the inherent mechanisms of different fusion tasks. Then, dual encoders map source images into a shared latent space, capturing common content while isolating modality-specific details. During the decoding phase, dual decoders act as feature injectors, selectively reintegrating the unique characteristics of each modality into the shared content for reconstruction. The modality-specific features are injected into the source image in the fusion process, generating the fused image that integrates information from both modalities. The architecture of decoders varies according to their fusion mechanisms, enhancing both performance and interpretability. Extensive experiments are conducted on diverse fusion tasks to validate the effectiveness and generalization ability of our method. The code is available at https://anonymous.4open.science/r/MAUGIF.
TiS-TSL: Image-Label Supervised Surgical Video Stereo Matching via Time-Switchable Teacher-Student Learning
Nov 12, 2025Abstract:Stereo matching in minimally invasive surgery (MIS) is essential for next-generation navigation and augmented reality. Yet, dense disparity supervision is nearly impossible due to anatomical constraints, typically limiting annotations to only a few image-level labels acquired before the endoscope enters deep body cavities. Teacher-Student Learning (TSL) offers a promising solution by leveraging a teacher trained on sparse labels to generate pseudo labels and associated confidence maps from abundant unlabeled surgical videos. However, existing TSL methods are confined to image-level supervision, providing only spatial confidence and lacking temporal consistency estimation. This absence of spatio-temporal reliability results in unstable disparity predictions and severe flickering artifacts across video frames. To overcome these challenges, we propose TiS-TSL, a novel time-switchable teacher-student learning framework for video stereo matching under minimal supervision. At its core is a unified model that operates in three distinct modes: Image-Prediction (IP), Forward Video-Prediction (FVP), and Backward Video-Prediction (BVP), enabling flexible temporal modeling within a single architecture. Enabled by this unified model, TiS-TSL adopts a two-stage learning strategy. The Image-to-Video (I2V) stage transfers sparse image-level knowledge to initialize temporal modeling. The subsequent Video-to-Video (V2V) stage refines temporal disparity predictions by comparing forward and backward predictions to calculate bidirectional spatio-temporal consistency. This consistency identifies unreliable regions across frames, filters noisy video-level pseudo labels, and enforces temporal coherence. Experimental results on two public datasets demonstrate that TiS-TSL exceeds other image-based state-of-the-arts by improving TEPE and EPE by at least 2.11% and 4.54%, respectively.
Bidirectional Mammogram View Translation with Column-Aware and Implicit 3D Conditional Diffusion
Oct 06, 2025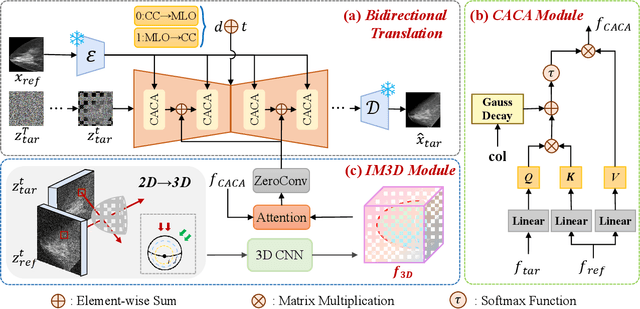
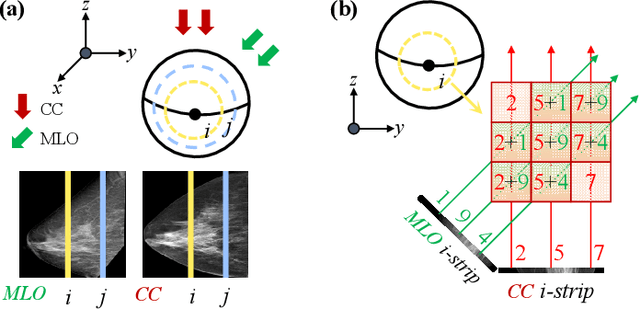
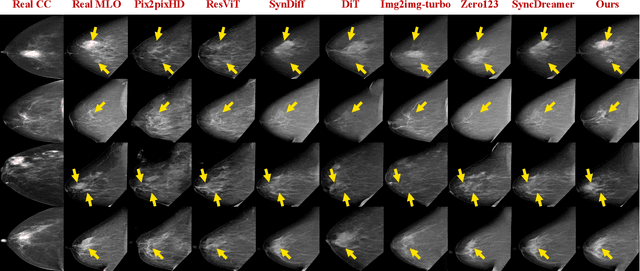
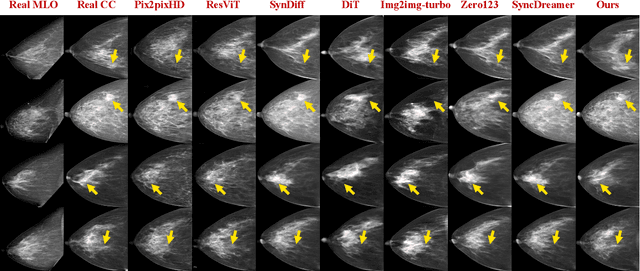
Abstract:Dual-view mammography, including craniocaudal (CC) and mediolateral oblique (MLO) projections, offers complementary anatomical views crucial for breast cancer diagnosis. However, in real-world clinical workflows, one view may be missing, corrupted, or degraded due to acquisition errors or compression artifacts, limiting the effectiveness of downstream analysis. View-to-view translation can help recover missing views and improve lesion alignment. Unlike natural images, this task in mammography is highly challenging due to large non-rigid deformations and severe tissue overlap in X-ray projections, which obscure pixel-level correspondences. In this paper, we propose Column-Aware and Implicit 3D Diffusion (CA3D-Diff), a novel bidirectional mammogram view translation framework based on conditional diffusion model. To address cross-view structural misalignment, we first design a column-aware cross-attention mechanism that leverages the geometric property that anatomically corresponding regions tend to lie in similar column positions across views. A Gaussian-decayed bias is applied to emphasize local column-wise correlations while suppressing distant mismatches. Furthermore, we introduce an implicit 3D structure reconstruction module that back-projects noisy 2D latents into a coarse 3D feature volume based on breast-view projection geometry. The reconstructed 3D structure is refined and injected into the denoising UNet to guide cross-view generation with enhanced anatomical awareness. Extensive experiments demonstrate that CA3D-Diff achieves superior performance in bidirectional tasks, outperforming state-of-the-art methods in visual fidelity and structural consistency. Furthermore, the synthesized views effectively improve single-view malignancy classification in screening settings, demonstrating the practical value of our method in real-world diagnostics.
FreeVPS: Repurposing Training-Free SAM2 for Generalizable Video Polyp Segmentation
Aug 27, 2025Abstract:Existing video polyp segmentation (VPS) paradigms usually struggle to balance between spatiotemporal modeling and domain generalization, limiting their applicability in real clinical scenarios. To embrace this challenge, we recast the VPS task as a track-by-detect paradigm that leverages the spatial contexts captured by the image polyp segmentation (IPS) model while integrating the temporal modeling capabilities of segment anything model 2 (SAM2). However, during long-term polyp tracking in colonoscopy videos, SAM2 suffers from error accumulation, resulting in a snowball effect that compromises segmentation stability. We mitigate this issue by repurposing SAM2 as a video polyp segmenter with two training-free modules. In particular, the intra-association filtering module eliminates spatial inaccuracies originating from the detecting stage, reducing false positives. The inter-association refinement module adaptively updates the memory bank to prevent error propagation over time, enhancing temporal coherence. Both modules work synergistically to stabilize SAM2, achieving cutting-edge performance in both in-domain and out-of-domain scenarios. Furthermore, we demonstrate the robust tracking capabilities of FreeVPS in long-untrimmed colonoscopy videos, underscoring its potential reliable clinical analysis.
Scalable Complexity Control Facilitates Reasoning Ability of LLMs
May 29, 2025



Abstract:The reasoning ability of large language models (LLMs) has been rapidly advancing in recent years, attracting interest in more fundamental approaches that can reliably enhance their generalizability. This work demonstrates that model complexity control, conveniently implementable by adjusting the initialization rate and weight decay coefficient, improves the scaling law of LLMs consistently over varying model sizes and data sizes. This gain is further illustrated by comparing the benchmark performance of 2.4B models pretrained on 1T tokens with different complexity hyperparameters. Instead of fixing the initialization std, we found that a constant initialization rate (the exponent of std) enables the scaling law to descend faster in both model and data sizes. These results indicate that complexity control is a promising direction for the continual advancement of LLMs.
Holistic White-light Polyp Classification via Alignment-free Dense Distillation of Auxiliary Optical Chromoendoscopy
May 25, 2025Abstract:White Light Imaging (WLI) and Narrow Band Imaging (NBI) are the two main colonoscopic modalities for polyp classification. While NBI, as optical chromoendoscopy, offers valuable vascular details, WLI remains the most common and often the only available modality in resource-limited settings. However, WLI-based methods typically underperform, limiting their clinical applicability. Existing approaches transfer knowledge from NBI to WLI through global feature alignment but often rely on cropped lesion regions, which are susceptible to detection errors and neglect contextual and subtle diagnostic cues. To address this, this paper proposes a novel holistic classification framework that leverages full-image diagnosis without requiring polyp localization. The key innovation lies in the Alignment-free Dense Distillation (ADD) module, which enables fine-grained cross-domain knowledge distillation regardless of misalignment between WLI and NBI images. Without resorting to explicit image alignment, ADD learns pixel-wise cross-domain affinities to establish correspondences between feature maps, guiding the distillation along the most relevant pixel connections. To further enhance distillation reliability, ADD incorporates Class Activation Mapping (CAM) to filter cross-domain affinities, ensuring the distillation path connects only those semantically consistent regions with equal contributions to polyp diagnosis. Extensive results on public and in-house datasets show that our method achieves state-of-the-art performance, relatively outperforming the other approaches by at least 2.5% and 16.2% in AUC, respectively. Code is available at: https://github.com/Huster-Hq/ADD.
KSHSeek: Data-Driven Approaches to Mitigating and Detecting Knowledge-Shortcut Hallucinations in Generative Models
Mar 25, 2025Abstract:The emergence of large language models (LLMs) has significantly advanced the development of natural language processing (NLP), especially in text generation tasks like question answering. However, model hallucinations remain a major challenge in natural language generation (NLG) tasks due to their complex causes. We systematically expand on the causes of factual hallucinations from the perspective of knowledge shortcuts, analyzing hallucinations arising from correct and defect-free data and demonstrating that knowledge-shortcut hallucinations are prevalent in generative models. To mitigate this issue, we propose a high similarity pruning algorithm at the data preprocessing level to reduce spurious correlations in the data. Additionally, we design a specific detection method for knowledge-shortcut hallucinations to evaluate the effectiveness of our mitigation strategy. Experimental results show that our approach effectively reduces knowledge-shortcut hallucinations, particularly in fine-tuning tasks, without negatively impacting model performance in question answering. This work introduces a new paradigm for mitigating specific hallucination issues in generative models, enhancing their robustness and reliability in real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge