Learning GFlowNets from partial episodes for improved convergence and stability
Sep 26, 2022Kanika Madan, Jarrid Rector-Brooks, Maksym Korablyov, Emmanuel Bengio, Moksh Jain, Andrei Nica, Tom Bosc, Yoshua Bengio, Nikolay Malkin
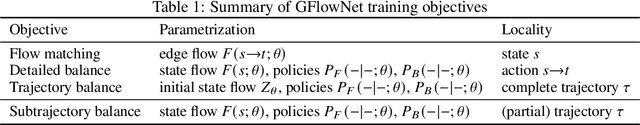

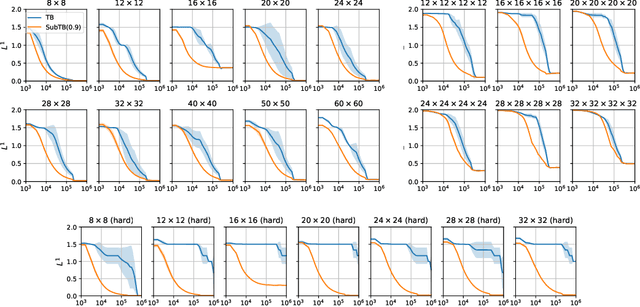
Generative flow networks (GFlowNets) are a family of algorithms for training a sequential sampler of discrete objects under an unnormalized target density and have been successfully used for various probabilistic modeling tasks. Existing training objectives for GFlowNets are either local to states or transitions, or propagate a reward signal over an entire sampling trajectory. We argue that these alternatives represent opposite ends of a gradient bias-variance tradeoff and propose a way to exploit this tradeoff to mitigate its harmful effects. Inspired by the TD($\lambda$) algorithm in reinforcement learning, we introduce subtrajectory balance or SubTB($\lambda$), a GFlowNet training objective that can learn from partial action subsequences of varying lengths. We show that SubTB($\lambda$) accelerates sampler convergence in previously studied and new environments and enables training GFlowNets in environments with longer action sequences and sparser reward landscapes than what was possible before. We also perform a comparative analysis of stochastic gradient dynamics, shedding light on the bias-variance tradeoff in GFlowNet training and the advantages of subtrajectory balance.
Interventional Causal Representation Learning
Sep 24, 2022Kartik Ahuja, Yixin Wang, Divyat Mahajan, Yoshua Bengio
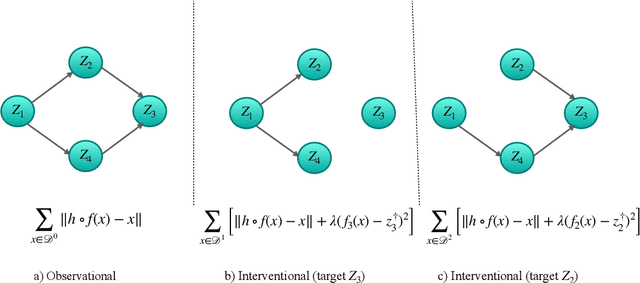
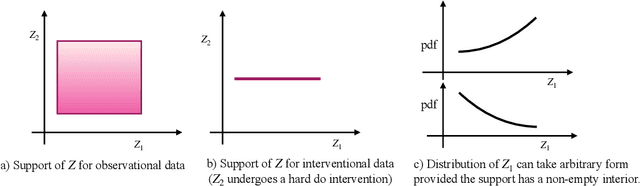
The theory of identifiable representation learning aims to build general-purpose methods that extract high-level latent (causal) factors from low-level sensory data. Most existing works focus on identifiable representation learning with observational data, relying on distributional assumptions on latent (causal) factors. However, in practice, we often also have access to interventional data for representation learning. How can we leverage interventional data to help identify high-level latents? To this end, we explore the role of interventional data for identifiable representation learning in this work. We study the identifiability of latent causal factors with and without interventional data, under minimal distributional assumptions on the latents. We prove that, if the true latent variables map to the observed high-dimensional data via a polynomial function, then representation learning via minimizing the standard reconstruction loss of autoencoders identifies the true latents up to affine transformation. If we further have access to interventional data generated by hard $do$ interventions on some of the latents, then we can identify these intervened latents up to permutation, shift and scaling.
Graph-Based Active Machine Learning Method for Diverse and Novel Antimicrobial Peptides Generation and Selection
Sep 18, 2022Bonaventure F. P. Dossou, Dianbo Liu, Xu Ji, Moksh Jain, Almer M. van der Sloot, Roger Palou, Michael Tyers, Yoshua Bengio
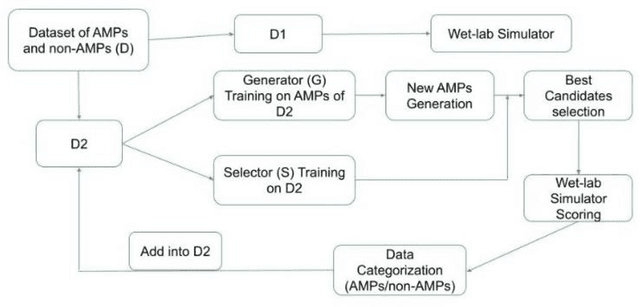
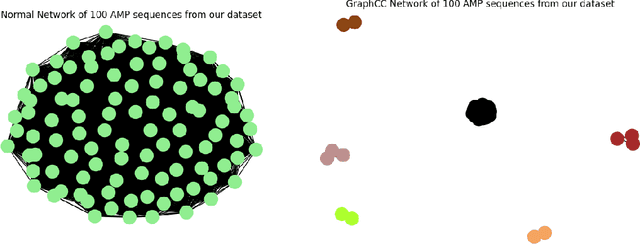
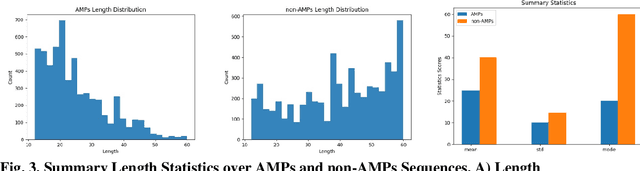
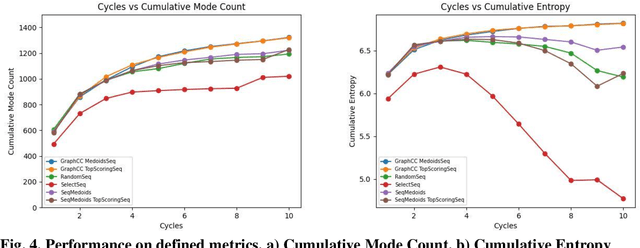
As antibiotic-resistant bacterial strains are rapidly spreading worldwide, infections caused by these strains are emerging as a global crisis causing the death of millions of people every year. Antimicrobial Peptides (AMPs) are one of the candidates to tackle this problem because of their potential diversity, and ability to favorably modulate the host immune response. However, large-scale screening of new AMP candidates is expensive, time-consuming, and now affordable in developing countries, which need the treatments the most. In this work, we propose a novel active machine learning-based framework that statistically minimizes the number of wet-lab experiments needed to design new AMPs, while ensuring a high diversity and novelty of generated AMPs sequences, in multi-rounds of wet-lab AMP screening settings. Combining recurrent neural network models and a graph-based filter (GraphCC), our proposed approach delivers novel and diverse candidates and demonstrates better performances according to our defined metrics.
Designing Biological Sequences via Meta-Reinforcement Learning and Bayesian Optimization
Sep 13, 2022Leo Feng, Padideh Nouri, Aneri Muni, Yoshua Bengio, Pierre-Luc Bacon
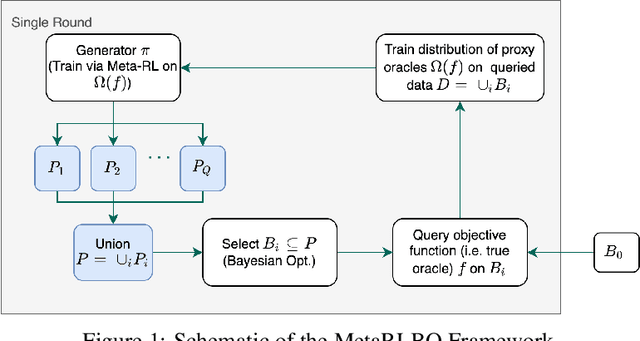
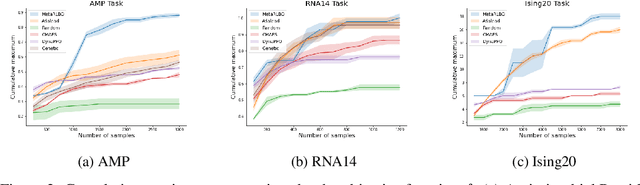
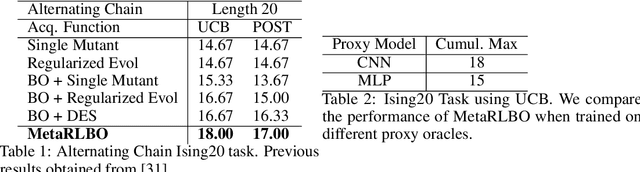
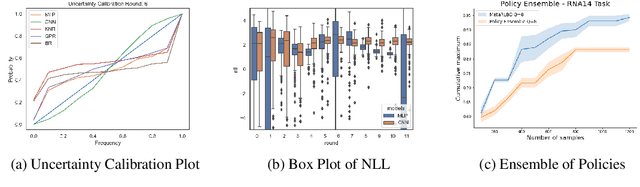
The ability to accelerate the design of biological sequences can have a substantial impact on the progress of the medical field. The problem can be framed as a global optimization problem where the objective is an expensive black-box function such that we can query large batches restricted with a limitation of a low number of rounds. Bayesian Optimization is a principled method for tackling this problem. However, the astronomically large state space of biological sequences renders brute-force iterating over all possible sequences infeasible. In this paper, we propose MetaRLBO where we train an autoregressive generative model via Meta-Reinforcement Learning to propose promising sequences for selection via Bayesian Optimization. We pose this problem as that of finding an optimal policy over a distribution of MDPs induced by sampling subsets of the data acquired in the previous rounds. Our in-silico experiments show that meta-learning over such ensembles provides robustness against reward misspecification and achieves competitive results compared to existing strong baselines.
Unifying Generative Models with GFlowNets
Sep 06, 2022Dinghuai Zhang, Ricky T. Q. Chen, Nikolay Malkin, Yoshua Bengio
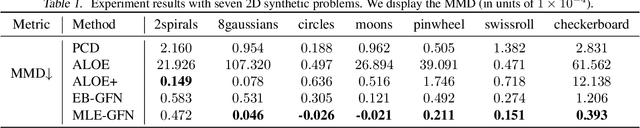
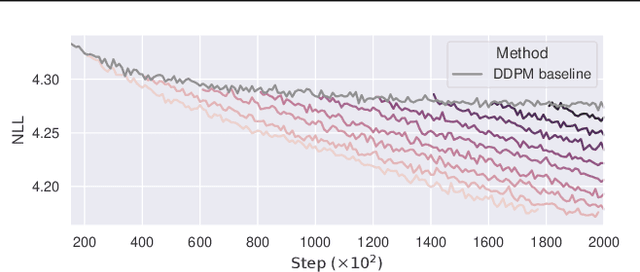
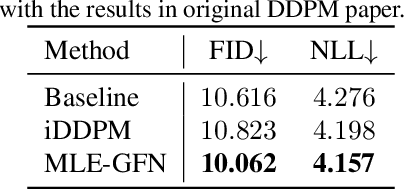
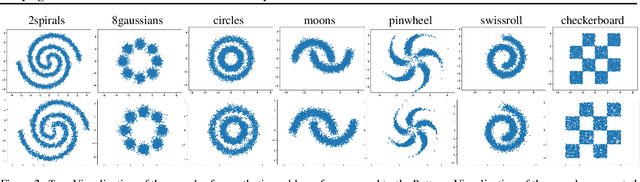
There are many frameworks for deep generative modeling, each often presented with their own specific training algorithms and inference methods. We present a short note on the connections between existing deep generative models and the GFlowNet framework, shedding light on their overlapping traits and providing a unifying viewpoint through the lens of learning with Markovian trajectories. This provides a means for unifying training and inference algorithms, and provides a route to construct an agglomeration of generative models.
AI for Global Climate Cooperation: Modeling Global Climate Negotiations, Agreements, and Long-Term Cooperation in RICE-N
Aug 15, 2022Tianyu Zhang, Andrew Williams, Soham Phade, Sunil Srinivasa, Yang Zhang, Prateek Gupta, Yoshua Bengio, Stephan Zheng
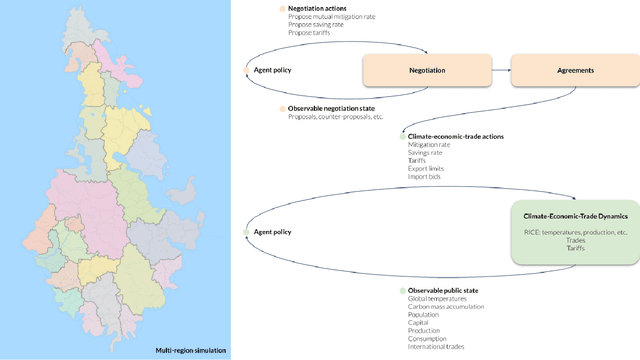
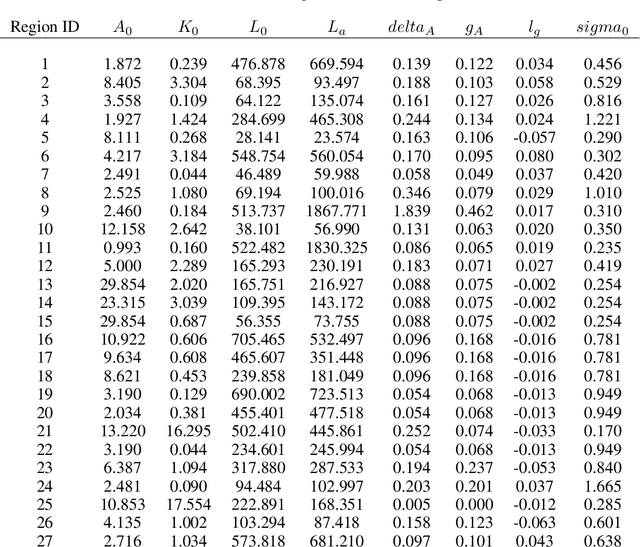
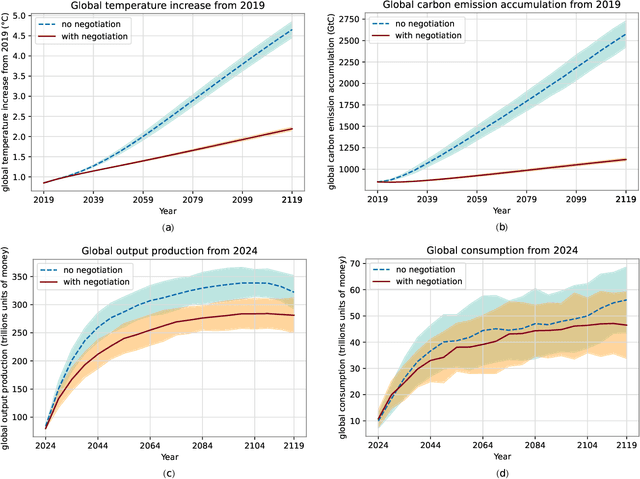
Comprehensive global cooperation is essential to limit global temperature increases while continuing economic development, e.g., reducing severe inequality or achieving long-term economic growth. Achieving long-term cooperation on climate change mitigation with n strategic agents poses a complex game-theoretic problem. For example, agents may negotiate and reach climate agreements, but there is no central authority to enforce adherence to those agreements. Hence, it is critical to design negotiation and agreement frameworks that foster cooperation, allow all agents to meet their individual policy objectives, and incentivize long-term adherence. This is an interdisciplinary challenge that calls for collaboration between researchers in machine learning, economics, climate science, law, policy, ethics, and other fields. In particular, we argue that machine learning is a critical tool to address the complexity of this domain. To facilitate this research, here we introduce RICE-N, a multi-region integrated assessment model that simulates the global climate and economy, and which can be used to design and evaluate the strategic outcomes for different negotiation and agreement frameworks. We also describe how to use multi-agent reinforcement learning to train rational agents using RICE-N. This framework underpinsAI for Global Climate Cooperation, a working group collaboration and competition on climate negotiation and agreement design. Here, we invite the scientific community to design and evaluate their solutions using RICE-N, machine learning, economic intuition, and other domain knowledge. More information can be found on www.ai4climatecoop.org.
Diversifying Design of Nucleic Acid Aptamers Using Unsupervised Machine Learning
Aug 10, 2022Siba Moussa, Michael Kilgour, Clara Jans, Alex Hernandez-Garcia, Miroslava Cuperlovic-Culf, Yoshua Bengio, Lena Simine
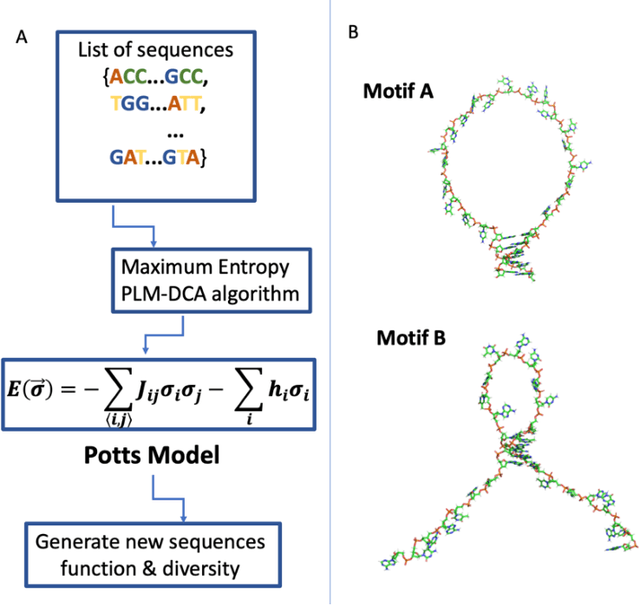
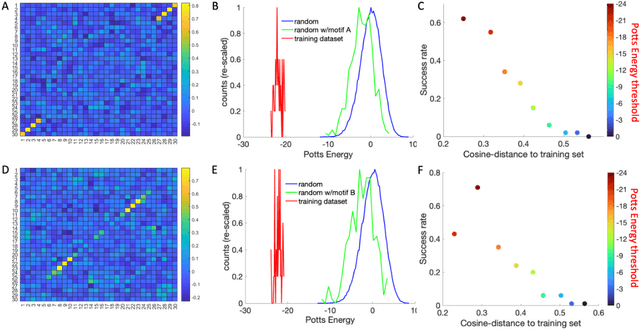
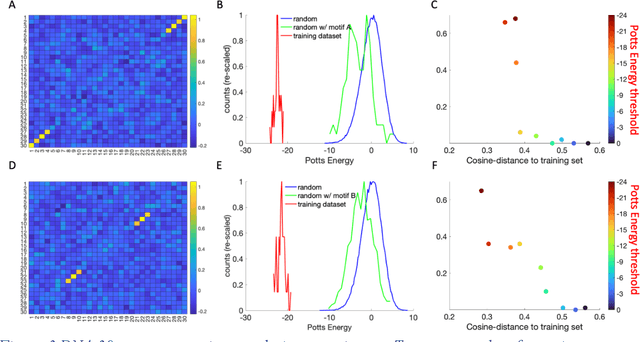
Inverse design of short single-stranded RNA and DNA sequences (aptamers) is the task of finding sequences that satisfy a set of desired criteria. Relevant criteria may be, for example, the presence of specific folding motifs, binding to molecular ligands, sensing properties, etc. Most practical approaches to aptamer design identify a small set of promising candidate sequences using high-throughput experiments (e.g. SELEX), and then optimize performance by introducing only minor modifications to the empirically found candidates. Sequences that possess the desired properties but differ drastically in chemical composition will add diversity to the search space and facilitate the discovery of useful nucleic acid aptamers. Systematic diversification protocols are needed. Here we propose to use an unsupervised machine learning model known as the Potts model to discover new, useful sequences with controllable sequence diversity. We start by training a Potts model using the maximum entropy principle on a small set of empirically identified sequences unified by a common feature. To generate new candidate sequences with a controllable degree of diversity, we take advantage of the model's spectral feature: an energy bandgap separating sequences that are similar to the training set from those that are distinct. By controlling the Potts energy range that is sampled, we generate sequences that are distinct from the training set yet still likely to have the encoded features. To demonstrate performance, we apply our approach to design diverse pools of sequences with specified secondary structure motifs in 30-mer RNA and DNA aptamers.
Controlled Sparsity via Constrained Optimization or: How I Learned to Stop Tuning Penalties and Love Constraints
Aug 08, 2022Jose Gallego-Posada, Juan Ramirez, Akram Erraqabi, Yoshua Bengio, Simon Lacoste-Julien
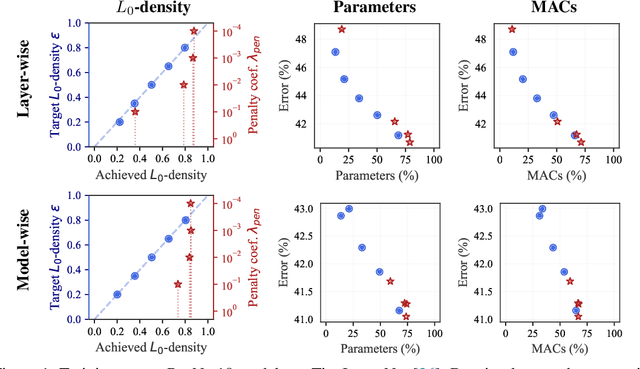
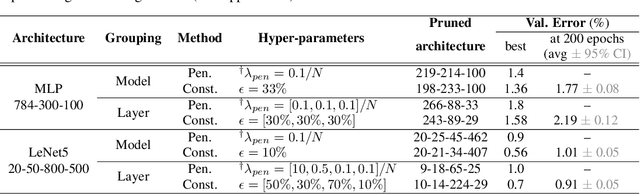
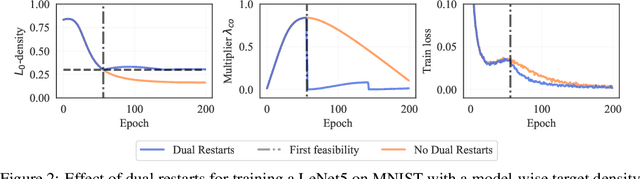
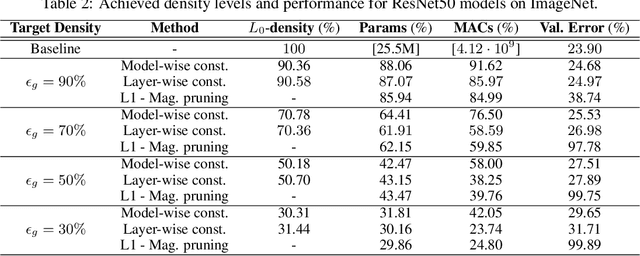
The performance of trained neural networks is robust to harsh levels of pruning. Coupled with the ever-growing size of deep learning models, this observation has motivated extensive research on learning sparse models. In this work, we focus on the task of controlling the level of sparsity when performing sparse learning. Existing methods based on sparsity-inducing penalties involve expensive trial-and-error tuning of the penalty factor, thus lacking direct control of the resulting model sparsity. In response, we adopt a constrained formulation: using the gate mechanism proposed by Louizos et al. (2018), we formulate a constrained optimization problem where sparsification is guided by the training objective and the desired sparsity target in an end-to-end fashion. Experiments on CIFAR-10/100, TinyImageNet, and ImageNet using WideResNet and ResNet{18, 50} models validate the effectiveness of our proposal and demonstrate that we can reliably achieve pre-determined sparsity targets without compromising on predictive performance.
Discrete Key-Value Bottleneck
Jul 22, 2022Frederik Träuble, Anirudh Goyal, Nasim Rahaman, Michael Mozer, Kenji Kawaguchi, Yoshua Bengio, Bernhard Schölkopf
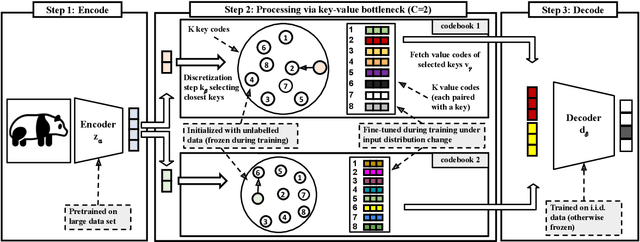
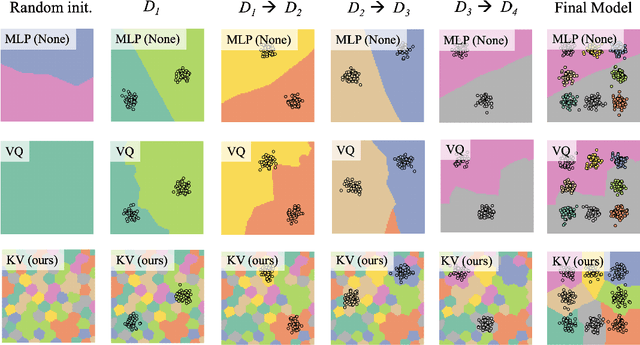
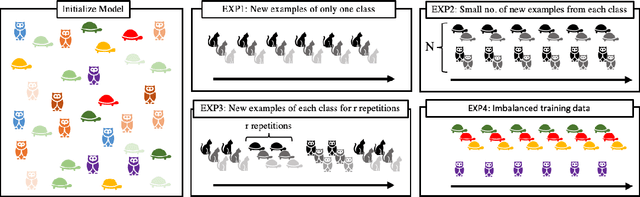
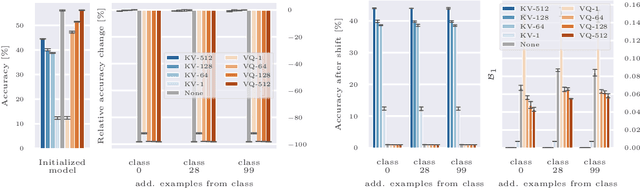
Deep neural networks perform well on prediction and classification tasks in the canonical setting where data streams are i.i.d., labeled data is abundant, and class labels are balanced. Challenges emerge with distribution shifts, including non-stationary or imbalanced data streams. One powerful approach that has addressed this challenge involves self-supervised pretraining of large encoders on volumes of unlabeled data, followed by task-specific tuning. Given a new task, updating the weights of these encoders is challenging as a large number of weights needs to be fine-tuned, and as a result, they forget information about the previous tasks. In the present work, we propose a model architecture to address this issue, building upon a discrete bottleneck containing pairs of separate and learnable (key, value) codes. In this setup, we follow the encode; process the representation via a discrete bottleneck; and decode paradigm, where the input is fed to the pretrained encoder, the output of the encoder is used to select the nearest keys, and the corresponding values are fed to the decoder to solve the current task. The model can only fetch and re-use a limited number of these (key, value) pairs during inference, enabling localized and context-dependent model updates. We theoretically investigate the ability of the proposed model to minimize the effect of the distribution shifts and show that such a discrete bottleneck with (key, value) pairs reduces the complexity of the hypothesis class. We empirically verified the proposed methods' benefits under challenging distribution shift scenarios across various benchmark datasets and show that the proposed model reduces the common vulnerability to non-i.i.d. and non-stationary training distributions compared to various other baselines.
Lookback for Learning to Branch
Jun 30, 2022Prateek Gupta, Elias B. Khalil, Didier Chetélat, Maxime Gasse, Yoshua Bengio, Andrea Lodi, M. Pawan Kumar
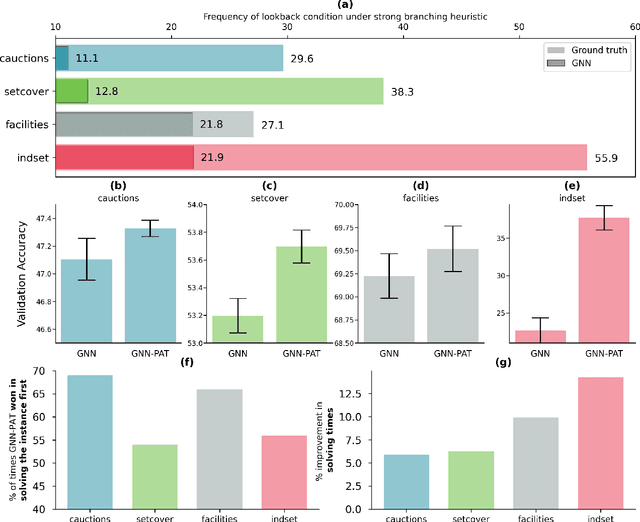

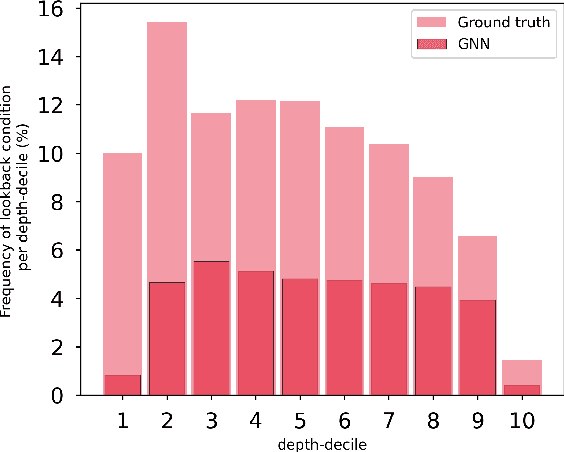
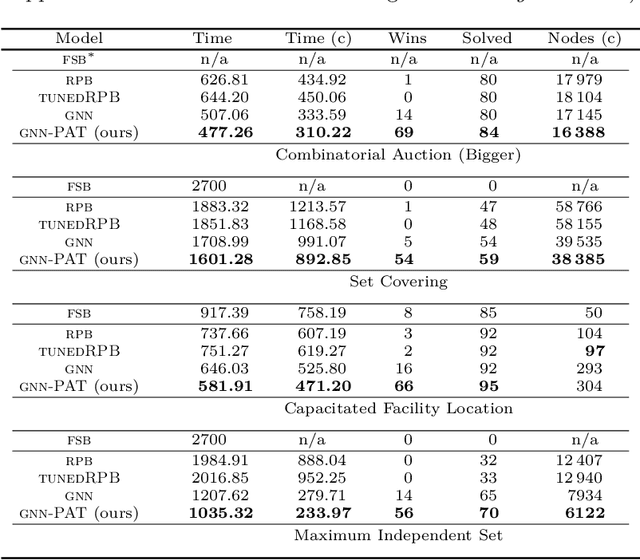
The expressive and computationally inexpensive bipartite Graph Neural Networks (GNN) have been shown to be an important component of deep learning based Mixed-Integer Linear Program (MILP) solvers. Recent works have demonstrated the effectiveness of such GNNs in replacing the branching (variable selection) heuristic in branch-and-bound (B&B) solvers. These GNNs are trained, offline and on a collection of MILPs, to imitate a very good but computationally expensive branching heuristic, strong branching. Given that B&B results in a tree of sub-MILPs, we ask (a) whether there are strong dependencies exhibited by the target heuristic among the neighboring nodes of the B&B tree, and (b) if so, whether we can incorporate them in our training procedure. Specifically, we find that with the strong branching heuristic, a child node's best choice was often the parent's second-best choice. We call this the "lookback" phenomenon. Surprisingly, the typical branching GNN of Gasse et al. (2019) often misses this simple "answer". To imitate the target behavior more closely by incorporating the lookback phenomenon in GNNs, we propose two methods: (a) target smoothing for the standard cross-entropy loss function, and (b) adding a Parent-as-Target (PAT) Lookback regularizer term. Finally, we propose a model selection framework to incorporate harder-to-formulate objectives such as solving time in the final models. Through extensive experimentation on standard benchmark instances, we show that our proposal results in up to 22% decrease in the size of the B&B tree and up to 15% improvement in the solving times.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge