Xueyu Liu
D4PM: A Dual-branch Driven Denoising Diffusion Probabilistic Model with Joint Posterior Diffusion Sampling for EEG Artifacts Removal
Sep 17, 2025Abstract:Artifact removal is critical for accurate analysis and interpretation of Electroencephalogram (EEG) signals. Traditional methods perform poorly with strong artifact-EEG correlations or single-channel data. Recent advances in diffusion-based generative models have demonstrated strong potential for EEG denoising, notably improving fine-grained noise suppression and reducing over-smoothing. However, existing methods face two main limitations: lack of temporal modeling limits interpretability and the use of single-artifact training paradigms ignore inter-artifact differences. To address these issues, we propose D4PM, a dual-branch driven denoising diffusion probabilistic model that unifies multi-type artifact removal. We introduce a dual-branch conditional diffusion architecture to implicitly model the data distribution of clean EEG and artifacts. A joint posterior sampling strategy is further designed to collaboratively integrate complementary priors for high-fidelity EEG reconstruction. Extensive experiments on two public datasets show that D4PM delivers superior denoising. It achieves new state-of-the-art performance in EOG artifact removal, outperforming all publicly available baselines. The code is available at https://github.com/flysnow1024/D4PM.
FMaMIL: Frequency-Driven Mamba Multi-Instance Learning for Weakly Supervised Lesion Segmentation in Medical Images
Jun 09, 2025Abstract:Accurate lesion segmentation in histopathology images is essential for diagnostic interpretation and quantitative analysis, yet it remains challenging due to the limited availability of costly pixel-level annotations. To address this, we propose FMaMIL, a novel two-stage framework for weakly supervised lesion segmentation based solely on image-level labels. In the first stage, a lightweight Mamba-based encoder is introduced to capture long-range dependencies across image patches under the MIL paradigm. To enhance spatial sensitivity and structural awareness, we design a learnable frequency-domain encoding module that supplements spatial-domain features with spectrum-based information. CAMs generated in this stage are used to guide segmentation training. In the second stage, we refine the initial pseudo labels via a CAM-guided soft-label supervision and a self-correction mechanism, enabling robust training even under label noise. Extensive experiments on both public and private histopathology datasets demonstrate that FMaMIL outperforms state-of-the-art weakly supervised methods without relying on pixel-level annotations, validating its effectiveness and potential for digital pathology applications.
DSAGL: Dual-Stream Attention-Guided Learning for Weakly Supervised Whole Slide Image Classification
May 29, 2025Abstract:Whole-slide images (WSIs) are critical for cancer diagnosis due to their ultra-high resolution and rich semantic content. However, their massive size and the limited availability of fine-grained annotations pose substantial challenges for conventional supervised learning. We propose DSAGL (Dual-Stream Attention-Guided Learning), a novel weakly supervised classification framework that combines a teacher-student architecture with a dual-stream design. DSAGL explicitly addresses instance-level ambiguity and bag-level semantic consistency by generating multi-scale attention-based pseudo labels and guiding instance-level learning. A shared lightweight encoder (VSSMamba) enables efficient long-range dependency modeling, while a fusion-attentive module (FASA) enhances focus on sparse but diagnostically relevant regions. We further introduce a hybrid loss to enforce mutual consistency between the two streams. Experiments on CIFAR-10, NCT-CRC, and TCGA-Lung datasets demonstrate that DSAGL consistently outperforms state-of-the-art MIL baselines, achieving superior discriminative performance and robustness under weak supervision.
Feature-prompting GBMSeg: One-Shot Reference Guided Training-Free Prompt Engineering for Glomerular Basement Membrane Segmentation
Jun 24, 2024



Abstract:Assessment of the glomerular basement membrane (GBM) in transmission electron microscopy (TEM) is crucial for diagnosing chronic kidney disease (CKD). The lack of domain-independent automatic segmentation tools for the GBM necessitates an AI-based solution to automate the process. In this study, we introduce GBMSeg, a training-free framework designed to automatically segment the GBM in TEM images guided only by a one-shot annotated reference. Specifically, GBMSeg first exploits the robust feature matching capabilities of the pretrained foundation model to generate initial prompt points, then introduces a series of novel automatic prompt engineering techniques across the feature and physical space to optimize the prompt scheme. Finally, GBMSeg employs a class-agnostic foundation segmentation model with the generated prompt scheme to obtain accurate segmentation results. Experimental results on our collected 2538 TEM images confirm that GBMSeg achieves superior segmentation performance with a Dice similarity coefficient (DSC) of 87.27% using only one labeled reference image in a training-free manner, outperforming recently proposed one-shot or few-shot methods. In summary, GBMSeg introduces a distinctive automatic prompt framework that facilitates robust domain-independent segmentation performance without training, particularly advancing the automatic prompting of foundation segmentation models for medical images. Future work involves automating the thickness measurement of segmented GBM and quantifying pathological indicators, holding significant potential for advancing pathology assessments in clinical applications. The source code is available on https://github.com/SnowRain510/GBMSeg
Graph-based multi-Feature fusion method for speech emotion recognition
Jun 11, 2024Abstract:Exploring proper way to conduct multi-speech feature fusion for cross-corpus speech emotion recognition is crucial as different speech features could provide complementary cues reflecting human emotion status. While most previous approaches only extract a single speech feature for emotion recognition, existing fusion methods such as concatenation, parallel connection, and splicing ignore heterogeneous patterns in the interaction between features and features, resulting in performance of existing systems. In this paper, we propose a novel graph-based fusion method to explicitly model the relationships between every pair of speech features. Specifically, we propose a multi-dimensional edge features learning strategy called Graph-based multi-Feature fusion method for speech emotion recognition. It represents each speech feature as a node and learns multi-dimensional edge features to explicitly describe the relationship between each feature-feature pair in the context of emotion recognition. This way, the learned multi-dimensional edge features encode speech feature-level information from both the vertex and edge dimensions. Our Approach consists of three modules: an Audio Feature Generation(AFG)module, an Audio-Feature Multi-dimensional Edge Feature(AMEF) module and a Speech Emotion Recognition (SER) module. The proposed methodology yielded satisfactory outcomes on the SEWA dataset. Furthermore, the method demonstrated enhanced performance compared to the baseline in the AVEC 2019 Workshop and Challenge. We used data from two cultures as our training and validation sets: two cultures containing German and Hungarian on the SEWA dataset, the CCC scores for German are improved by 17.28% for arousal and 7.93% for liking. The outcomes of our methodology demonstrate a 13% improvement over alternative fusion techniques, including those employing one dimensional edge-based feature fusion approach.
MSA-MIL: A deep residual multiple instance learning model based on multi-scale annotation for classification and visualization of glomerular spikes
Jul 02, 2020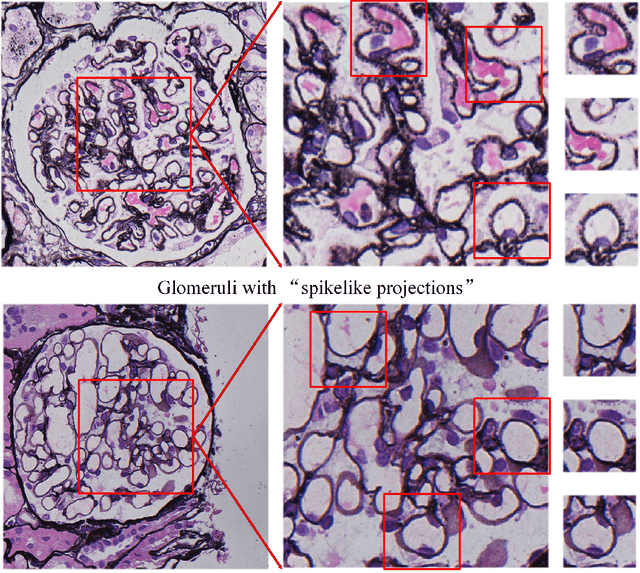
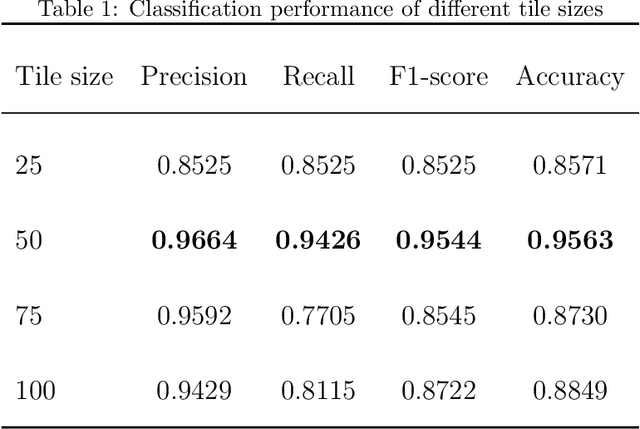
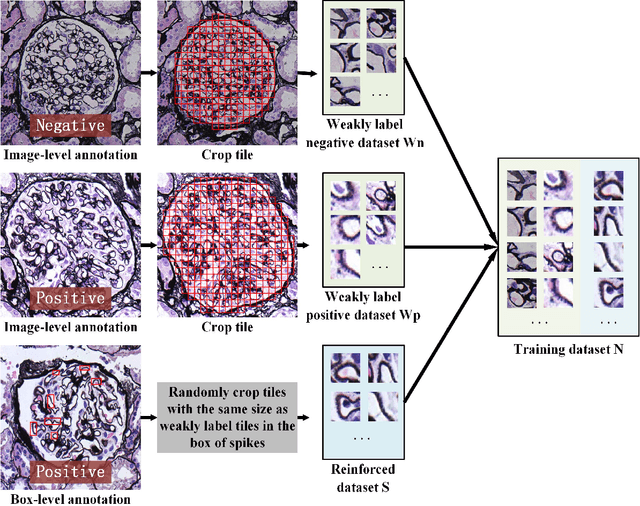
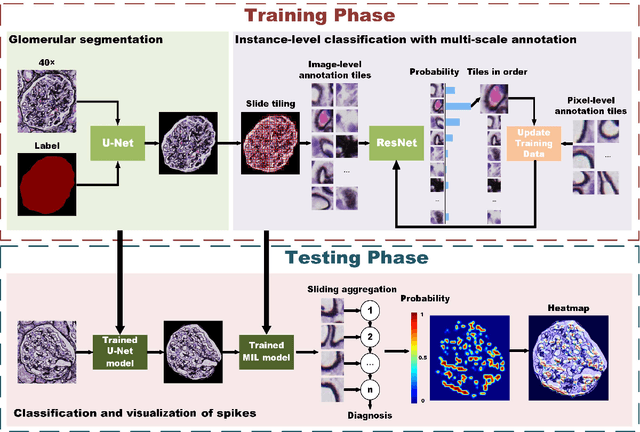
Abstract:Membranous nephropathy (MN) is a frequent type of adult nephrotic syndrome, which has a high clinical incidence and can cause various complications. In the biopsy microscope slide of membranous nephropathy, spikelike projections on the glomerular basement membrane is a prominent feature of the MN. However, due to the whole biopsy slide contains large number of glomeruli, and each glomerulus includes many spike lesions, the pathological feature of the spikes is not obvious. It thus is time-consuming for doctors to diagnose glomerulus one by one and is difficult for pathologists with less experience to diagnose. In this paper, we establish a visualized classification model based on the multi-scale annotation multi-instance learning (MSA-MIL) to achieve glomerular classification and spikes visualization. The MSA-MIL model mainly involves three parts. Firstly, U-Net is used to extract the region of the glomeruli to ensure that the features learned by the succeeding algorithm are focused inside the glomeruli itself. Secondly, we use MIL to train an instance-level classifier combined with MSA method to enhance the learning ability of the network by adding a location-level labeled reinforced dataset, thereby obtaining an example-level feature representation with rich semantics. Lastly, the predicted scores of each tile in the image are summarized to obtain glomerular classification and visualization of the classification results of the spikes via the usage of sliding window method. The experimental results confirm that the proposed MSA-MIL model can effectively and accurately classify normal glomeruli and spiked glomerulus and visualize the position of spikes in the glomerulus. Therefore, the proposed model can provide a good foundation for assisting the clinical doctors to diagnose the glomerular membranous nephropathy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge