Xavier Lladó
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025



Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024



Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
The state-of-the-art 3D anisotropic intracranial hemorrhage segmentation on non-contrast head CT: The INSTANCE challenge
Jan 12, 2023Abstract:Automatic intracranial hemorrhage segmentation in 3D non-contrast head CT (NCCT) scans is significant in clinical practice. Existing hemorrhage segmentation methods usually ignores the anisotropic nature of the NCCT, and are evaluated on different in-house datasets with distinct metrics, making it highly challenging to improve segmentation performance and perform objective comparisons among different methods. The INSTANCE 2022 was a grand challenge held in conjunction with the 2022 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). It is intended to resolve the above-mentioned problems and promote the development of both intracranial hemorrhage segmentation and anisotropic data processing. The INSTANCE released a training set of 100 cases with ground-truth and a validation set with 30 cases without ground-truth labels that were available to the participants. A held-out testing set with 70 cases is utilized for the final evaluation and ranking. The methods from different participants are ranked based on four metrics, including Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), Relative Volume Difference (RVD) and Normalized Surface Dice (NSD). A total of 13 teams submitted distinct solutions to resolve the challenges, making several baseline models, pre-processing strategies and anisotropic data processing techniques available to future researchers. The winner method achieved an average DSC of 0.6925, demonstrating a significant growth over our proposed baseline method. To the best of our knowledge, the proposed INSTANCE challenge releases the first intracranial hemorrhage segmentation benchmark, and is also the first challenge that intended to resolve the anisotropic problem in 3D medical image segmentation, which provides new alternatives in these research fields.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019
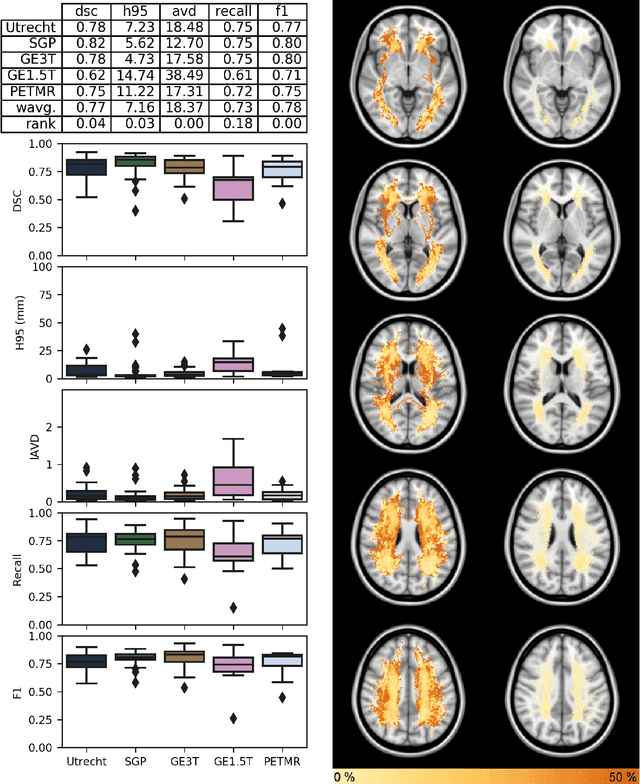
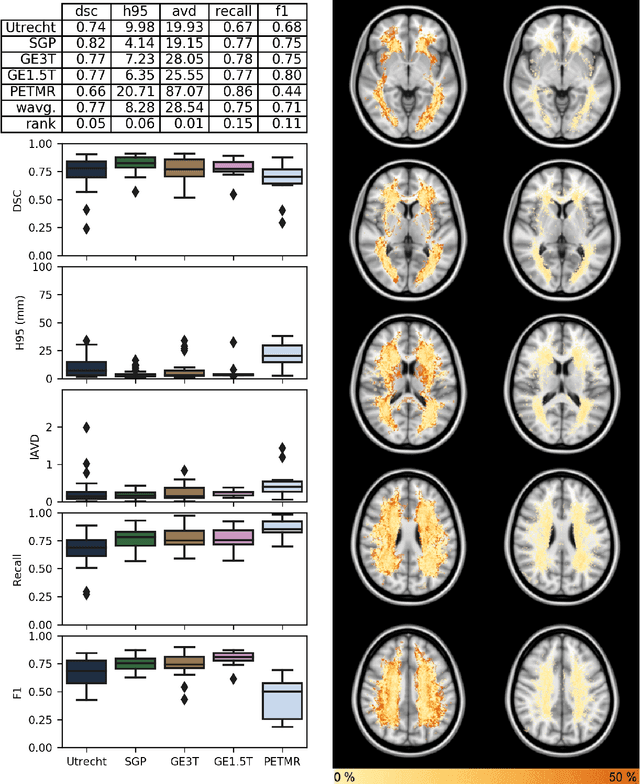
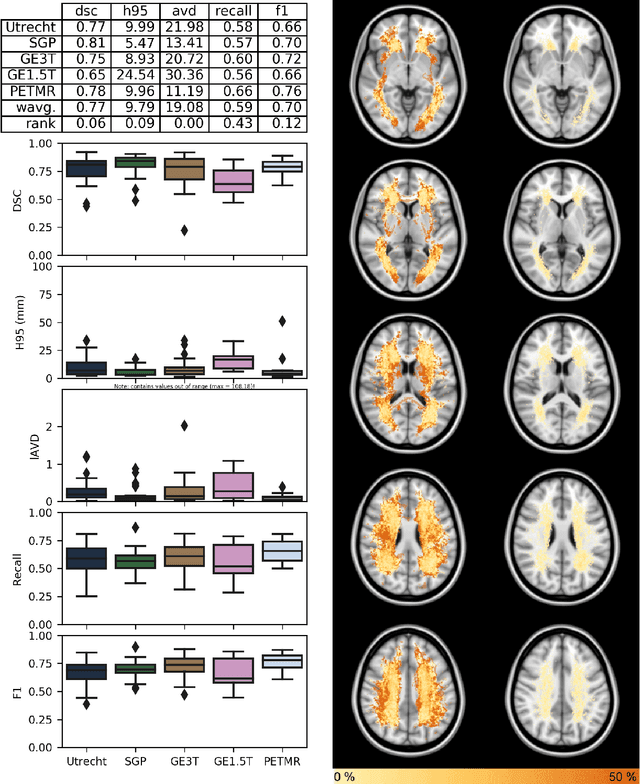
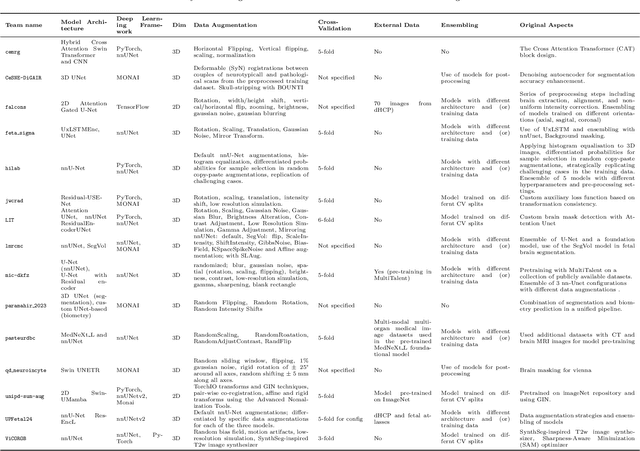
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Multiple Sclerosis Lesion Synthesis in MRI using an encoder-decoder U-NET
Jan 17, 2019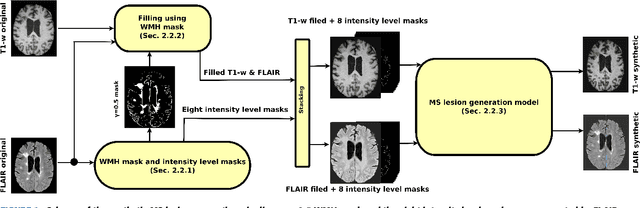
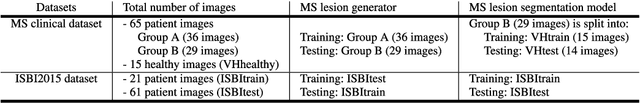

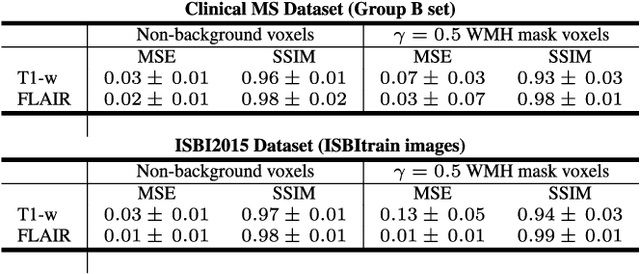
Abstract:In this paper, we propose generating synthetic multiple sclerosis (MS) lesions on MRI images with the final aim to improve the performance of supervised machine learning algorithms, therefore avoiding the problem of the lack of available ground truth. We propose a two-input two-output fully convolutional neural network model for MS lesion synthesis in MRI images. The lesion information is encoded as discrete binary intensity level masks passed to the model and stacked with the input images. The model is trained end-to-end without the need for manually annotating the lesions in the training set. We then perform the generation of synthetic lesions on healthy images via registration of patient images, which are subsequently used for data augmentation to increase the performance for supervised MS lesion detection algorithms. Our pipeline is evaluated on MS patient data from an in-house clinical dataset and the public ISBI2015 challenge dataset. The evaluation is based on measuring the similarities between the real and the synthetic images as well as in terms of lesion detection performance by segmenting both the original and synthetic images individually using a state-of-the-art segmentation framework. We also demonstrate the usage of synthetic MS lesions generated on healthy images as data augmentation. We analyze a scenario of limited training data (one-image training) to demonstrate the effect of the data augmentation on both datasets. Our results significantly show the effectiveness of the usage of synthetic MS lesion images. For the ISBI2015 challenge, our one-image model trained using only a single image plus the synthetic data augmentation strategy showed a performance similar to that of other CNN methods that were fully trained using the entire training set, yielding a comparable human expert rater performance
SUNet: a deep learning architecture for acute stroke lesion segmentation and outcome prediction in multimodal MRI
Oct 31, 2018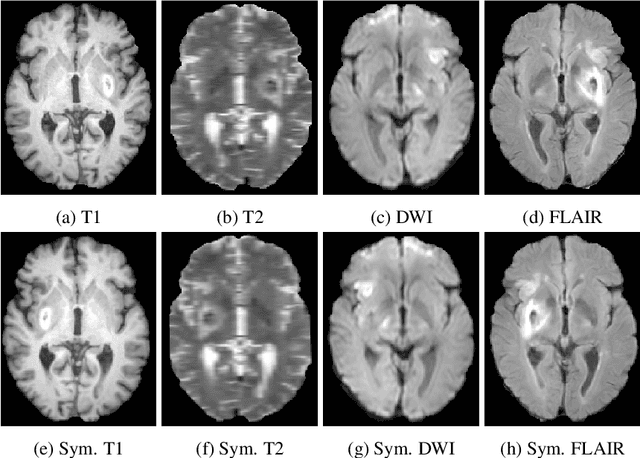
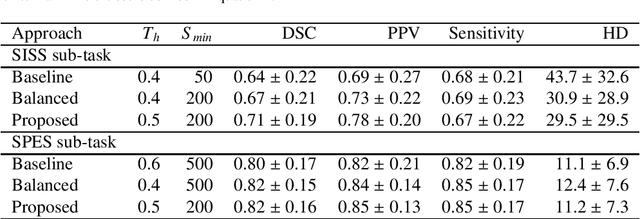
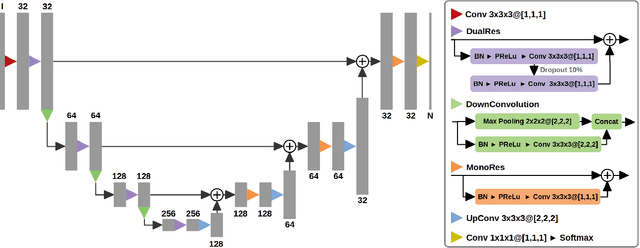
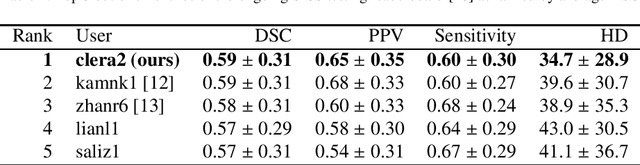
Abstract:Acute stroke lesion segmentation and prediction tasks are of great clinical interest as they can help doctors make better informed time-critical treatment decisions. Automatic segmentation of these lesions is a complex task due to their heterogeneous appearance, dynamic evolution and inter-patient differences. Typically, acute stroke lesion tasks are approached with methods developed for chronic stroke or other brain lesions. However, the pathophysiology and anatomy of acute stroke establishes an inherently different problem that needs special consideration. In this work, we propose a novel deep learning architecture specially designed for acute stroke tasks that involve approximating complex non-linear functions with reduced data. Within our strategy, class imbalance is tackled using a hybrid strategy based on state-of-the-art train sampling strategies designed for other brain lesion related tasks, which is more suited to the anatomy and pathophysiology of acute stroke lesions. The proposed method is evaluated on three unrelated public international challenge datasets (ISLES) without any dataset specific hyper-parameter tuning. These involve the tasks of sub-acute stroke lesion segmentation, acute stroke penumbra estimation and chronic extent prediction from acute MR images. The performance of the proposed architecture is analysed both against similar deep learning architectures from chronic stroke and related biomedical tasks and also by submitting the segmented test images for blind online evaluation on each of the challenges. When compared with the rest of submitted strategies, our method achieves top-rank performance among the best submitted entries in all the three challenges, showing its capability to deal with different unrelated tasks without hyper-parameter tuning. In order to promote the reproducibility of our results, a public version of the proposed method has been released.
Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review
Jun 11, 2018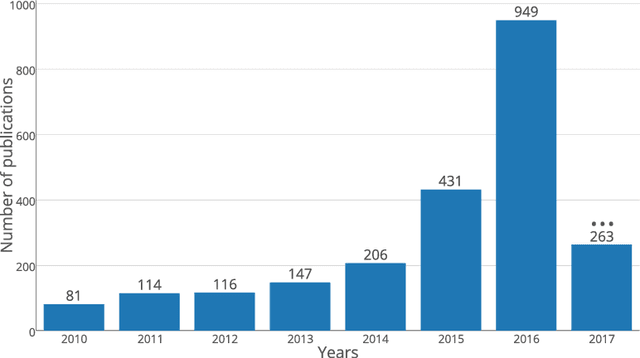
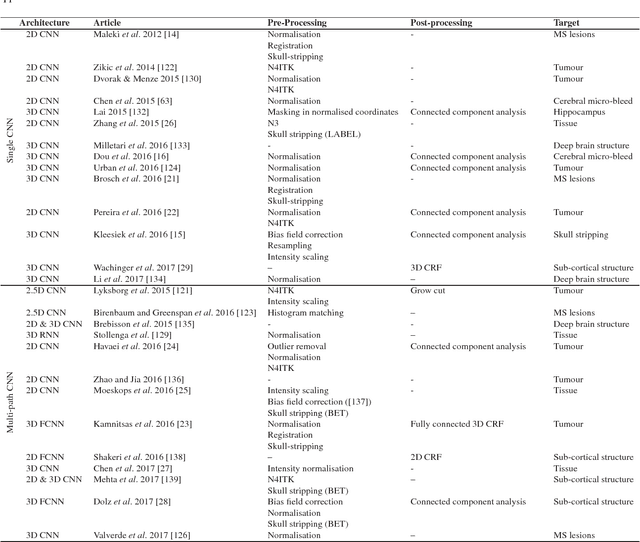
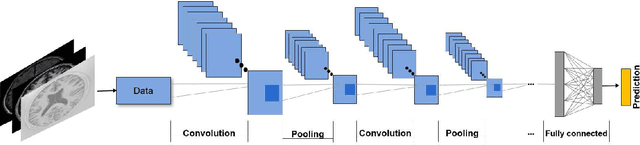
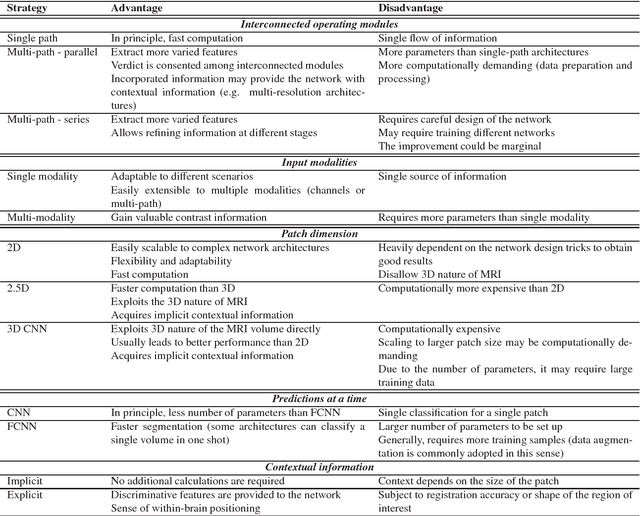
Abstract:In recent years, deep convolutional neural networks (CNNs) have shown record-shattering performance in a variety of computer vision problems, such as visual object recognition, detection and segmentation. These methods have also been utilised in medical image analysis domain for lesion segmentation, anatomical segmentation and classification. We present an extensive literature review of CNN techniques applied in brain magnetic resonance imaging (MRI) analysis, focusing on the architectures, pre-processing, data-preparation and post-processing strategies available in these works. The aim of this study is three-fold. Our primary goal is to report how different CNN architectures have evolved, discuss state-of-the-art strategies, condense their results obtained using public datasets and examine their pros and cons. Second, this paper is intended to be a detailed reference of the research activity in deep CNN for brain MRI analysis. Finally, we present a perspective on the future of CNNs in which we hint some of the research directions in subsequent years.
One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks
May 31, 2018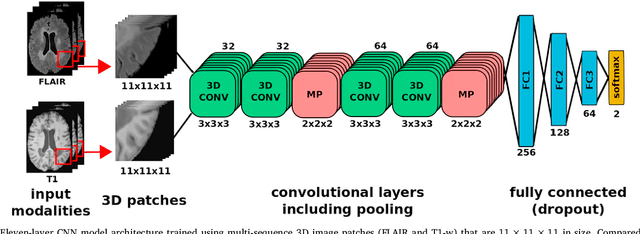
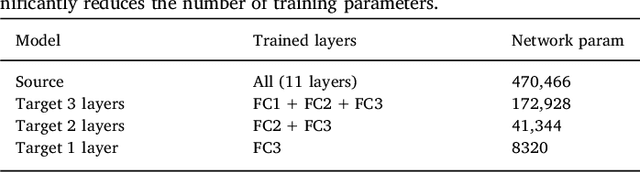
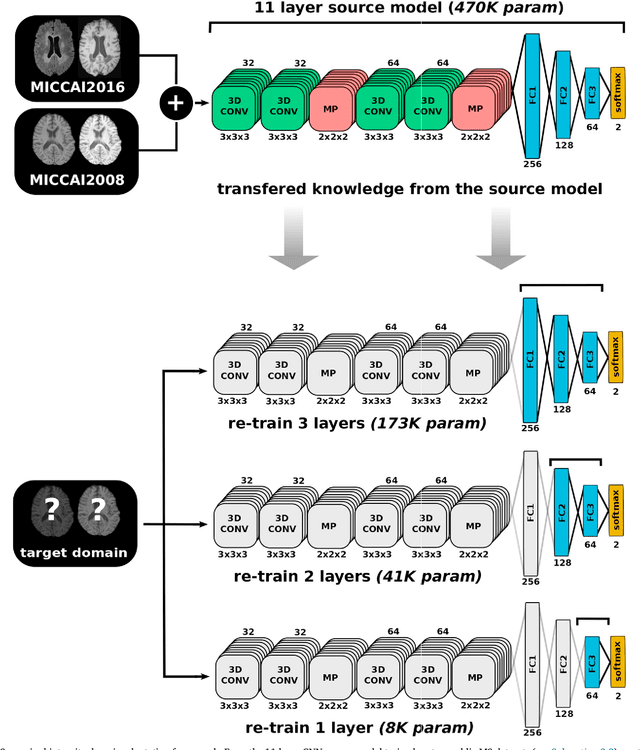
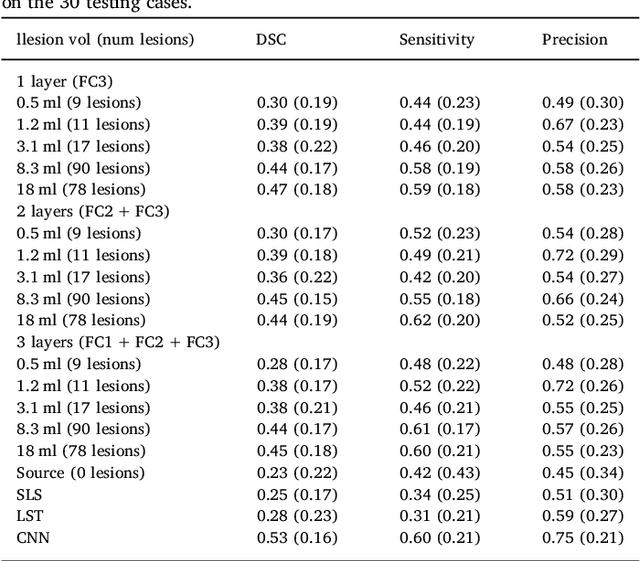
Abstract:In recent years, several convolutional neural network (CNN) methods have been proposed for the automated white matter lesion segmentation of multiple sclerosis (MS) patient images, due to their superior performance compared with those of other state-of-the-art methods. However, the accuracies of CNN methods tend to decrease significantly when evaluated on different image domains compared with those used for training, which demonstrates the lack of adaptability of CNNs to unseen imaging data. In this study, we analyzed the effect of intensity domain adaptation on our recently proposed CNN-based MS lesion segmentation method. Given a source model trained on two public MS datasets, we investigated the transferability of the CNN model when applied to other MRI scanners and protocols, evaluating the minimum number of annotated images needed from the new domain and the minimum number of layers needed to re-train to obtain comparable accuracy. Our analysis comprised MS patient data from both a clinical center and the public ISBI2015 challenge database, which permitted us to compare the domain adaptation capability of our model to that of other state-of-the-art methods. For the ISBI2015 challenge, our one-shot domain adaptation model trained using only a single image showed a performance similar to that of other CNN methods that were fully trained using the entire available training set, yielding a comparable human expert rater performance. We believe that our experiments will encourage the MS community to incorporate its use in different clinical settings with reduced amounts of annotated data. This approach could be meaningful not only in terms of the accuracy in delineating MS lesions but also in the related reductions in time and economic costs derived from manual lesion labeling.
Quantitative analysis of patch-based fully convolutional neural networks for tissue segmentation on brain magnetic resonance imaging
Feb 19, 2018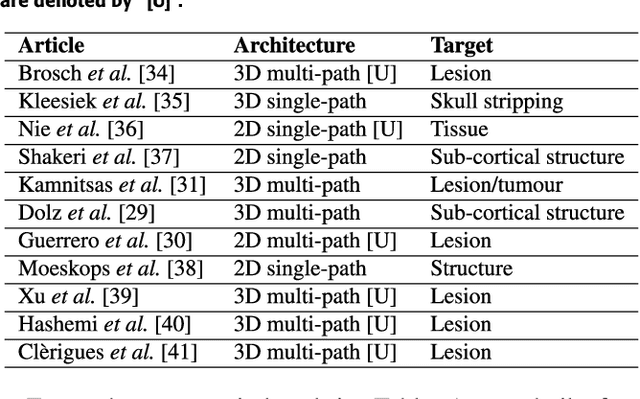
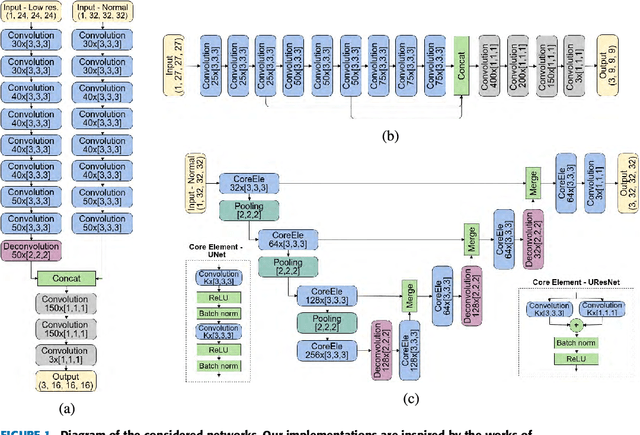
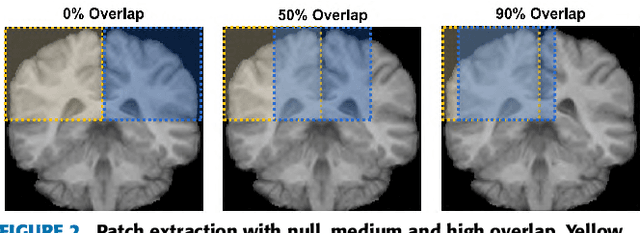
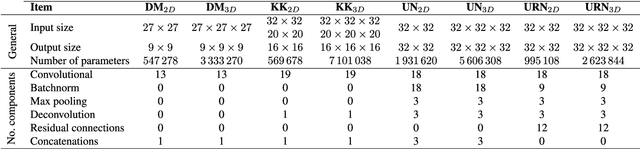
Abstract:Accurate brain tissue segmentation in Magnetic Resonance Imaging (MRI) has attracted the attention of medical doctors and researchers since variations in tissue volume help in diagnosing and monitoring neurological diseases. Several proposals have been designed throughout the years comprising conventional machine learning strategies as well as convolutional neural networks (CNN) approaches. In particular, in this paper, we analyse a sub-group of deep learning methods producing dense predictions. This branch, referred in the literature as Fully CNN (FCNN), is of interest as these architectures can process an input volume in less time than CNNs and local spatial dependencies may be encoded since several voxels are classified at once. Our study focuses on understanding architectural strengths and weaknesses of literature-like approaches. Hence, we implement eight FCNN architectures inspired by robust state-of-the-art methods on brain segmentation related tasks. We evaluate them using the IBSR18, MICCAI2012 and iSeg2017 datasets as they contain infant and adult data and exhibit varied voxel spacing, image quality, number of scans and available imaging modalities. The discussion is driven in three directions: comparison between 2D and 3D approaches, the importance of multiple modalities and overlapping as a sampling strategy for training and testing models. To encourage other researchers to explore the evaluation framework, a public version is accessible to download from our research website.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge