Zijiao Chen
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025



Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Prompt Your Brain: Scaffold Prompt Tuning for Efficient Adaptation of fMRI Pre-trained Model
Aug 20, 2024



Abstract:We introduce Scaffold Prompt Tuning (ScaPT), a novel prompt-based framework for adapting large-scale functional magnetic resonance imaging (fMRI) pre-trained models to downstream tasks, with high parameter efficiency and improved performance compared to fine-tuning and baselines for prompt tuning. The full fine-tuning updates all pre-trained parameters, which may distort the learned feature space and lead to overfitting with limited training data which is common in fMRI fields. In contrast, we design a hierarchical prompt structure that transfers the knowledge learned from high-resource tasks to low-resource ones. This structure, equipped with a Deeply-conditioned Input-Prompt (DIP) mapping module, allows for efficient adaptation by updating only 2% of the trainable parameters. The framework enhances semantic interpretability through attention mechanisms between inputs and prompts, and it clusters prompts in the latent space in alignment with prior knowledge. Experiments on public resting state fMRI datasets reveal ScaPT outperforms fine-tuning and multitask-based prompt tuning in neurodegenerative diseases diagnosis/prognosis and personality trait prediction, even with fewer than 20 participants. It highlights ScaPT's efficiency in adapting pre-trained fMRI models to low-resource tasks.
NeuroCine: Decoding Vivid Video Sequences from Human Brain Activties
Feb 02, 2024



Abstract:In the pursuit to understand the intricacies of human brain's visual processing, reconstructing dynamic visual experiences from brain activities emerges as a challenging yet fascinating endeavor. While recent advancements have achieved success in reconstructing static images from non-invasive brain recordings, the domain of translating continuous brain activities into video format remains underexplored. In this work, we introduce NeuroCine, a novel dual-phase framework to targeting the inherent challenges of decoding fMRI data, such as noises, spatial redundancy and temporal lags. This framework proposes spatial masking and temporal interpolation-based augmentation for contrastive learning fMRI representations and a diffusion model enhanced by dependent prior noise for video generation. Tested on a publicly available fMRI dataset, our method shows promising results, outperforming the previous state-of-the-art models by a notable margin of ${20.97\%}$, ${31.00\%}$ and ${12.30\%}$ respectively on decoding the brain activities of three subjects in the fMRI dataset, as measured by SSIM. Additionally, our attention analysis suggests that the model aligns with existing brain structures and functions, indicating its biological plausibility and interpretability.
Contrast, Attend and Diffuse to Decode High-Resolution Images from Brain Activities
May 26, 2023



Abstract:Decoding visual stimuli from neural responses recorded by functional Magnetic Resonance Imaging (fMRI) presents an intriguing intersection between cognitive neuroscience and machine learning, promising advancements in understanding human visual perception and building non-invasive brain-machine interfaces. However, the task is challenging due to the noisy nature of fMRI signals and the intricate pattern of brain visual representations. To mitigate these challenges, we introduce a two-phase fMRI representation learning framework. The first phase pre-trains an fMRI feature learner with a proposed Double-contrastive Mask Auto-encoder to learn denoised representations. The second phase tunes the feature learner to attend to neural activation patterns most informative for visual reconstruction with guidance from an image auto-encoder. The optimized fMRI feature learner then conditions a latent diffusion model to reconstruct image stimuli from brain activities. Experimental results demonstrate our model's superiority in generating high-resolution and semantically accurate images, substantially exceeding previous state-of-the-art methods by 39.34% in the 50-way-top-1 semantic classification accuracy. Our research invites further exploration of the decoding task's potential and contributes to the development of non-invasive brain-machine interfaces.
Cinematic Mindscapes: High-quality Video Reconstruction from Brain Activity
May 19, 2023



Abstract:Reconstructing human vision from brain activities has been an appealing task that helps to understand our cognitive process. Even though recent research has seen great success in reconstructing static images from non-invasive brain recordings, work on recovering continuous visual experiences in the form of videos is limited. In this work, we propose Mind-Video that learns spatiotemporal information from continuous fMRI data of the cerebral cortex progressively through masked brain modeling, multimodal contrastive learning with spatiotemporal attention, and co-training with an augmented Stable Diffusion model that incorporates network temporal inflation. We show that high-quality videos of arbitrary frame rates can be reconstructed with Mind-Video using adversarial guidance. The recovered videos were evaluated with various semantic and pixel-level metrics. We achieved an average accuracy of 85% in semantic classification tasks and 0.19 in structural similarity index (SSIM), outperforming the previous state-of-the-art by 45%. We also show that our model is biologically plausible and interpretable, reflecting established physiological processes.
Seeing Beyond the Brain: Conditional Diffusion Model with Sparse Masked Modeling for Vision Decoding
Nov 15, 2022



Abstract:Decoding visual stimuli from brain recordings aims to deepen our understanding of the human visual system and build a solid foundation for bridging human and computer vision through the Brain-Computer Interface. However, reconstructing high-quality images with correct semantics from brain recordings is a challenging problem due to the complex underlying representations of brain signals and the scarcity of data annotations. In this work, we present MinD-Vis: Sparse Masked Brain Modeling with Double-Conditioned Latent Diffusion Model for Human Vision Decoding. Firstly, we learn an effective self-supervised representation of fMRI data using mask modeling in a large latent space inspired by the sparse coding of information in the primary visual cortex. Then by augmenting a latent diffusion model with double-conditioning, we show that MinD-Vis can reconstruct highly plausible images with semantically matching details from brain recordings using very few paired annotations. We benchmarked our model qualitatively and quantitatively; the experimental results indicate that our method outperformed state-of-the-art in both semantic mapping (100-way semantic classification) and generation quality (FID) by 66% and 41% respectively. An exhaustive ablation study was also conducted to analyze our framework.
A Wearable ECG Monitor for Deep Learning Based Real-Time Cardiovascular Disease Detection
Jan 25, 2022
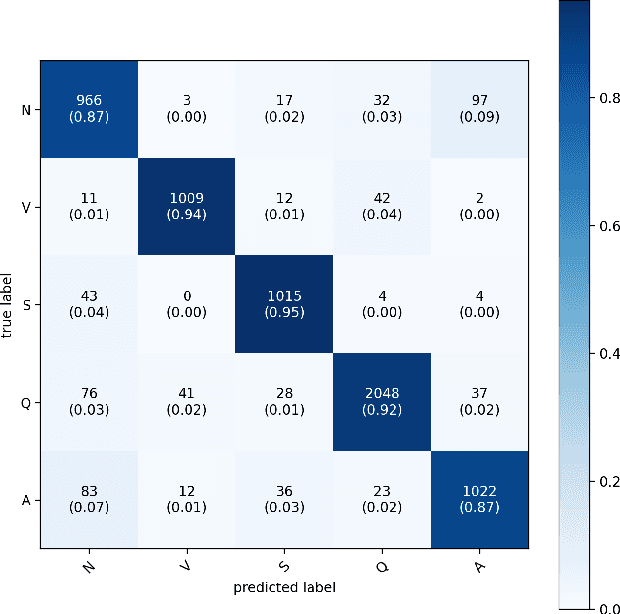

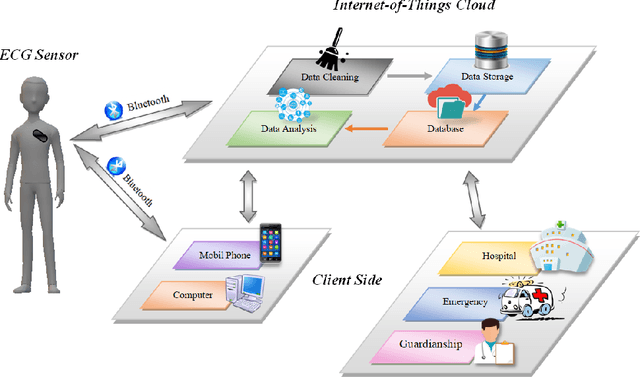
Abstract:Cardiovascular disease has become one of the most significant threats endangering human life and health. Recently, Electrocardiogram (ECG) monitoring has been transformed into remote cardiac monitoring by Holter surveillance. However, the widely used Holter can bring a great deal of discomfort and inconvenience to the individuals who carry them. We developed a new wireless ECG patch in this work and applied a deep learning framework based on the Convolutional Neural Network (CNN) and Long Short-term Memory (LSTM) models. However, we find that the models using the existing techniques are not able to differentiate two main heartbeat types (Supraventricular premature beat and Atrial fibrillation) in our newly obtained dataset, resulting in low accuracy of 58.0 %. We proposed a semi-supervised method to process the badly labelled data samples with using the confidence-level-based training. The experiment results conclude that the proposed method can approach an average accuracy of 90.2 %, i.e., 5.4 % higher than the accuracy of conventional ECG classification methods.
Negative-ResNet: Noisy Ambulatory Electrocardiogram Signal Classification Scheme
Jan 25, 2022
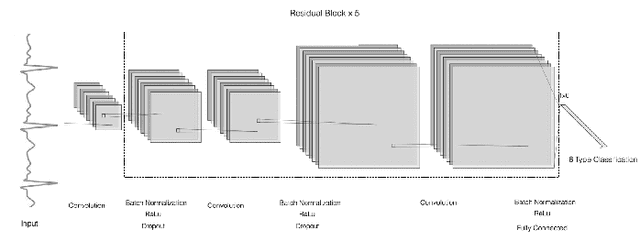
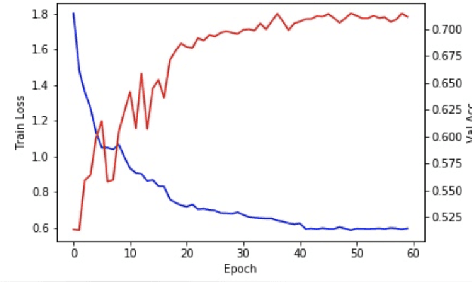
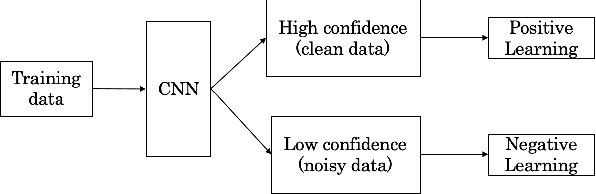
Abstract:With recently successful applications of deep learning in computer vision and general signal processing, deep learning has shown many unique advantages in medical signal processing. However, data labelling quality has become one of the most significant issues for AI applications, especially when it requires domain knowledge (e.g. medical image labelling). In addition, there might be noisy labels in practical datasets, which might impair the training process of neural networks. In this work, we propose a semi-supervised algorithm for training data samples with noisy labels by performing selected Positive Learning (PL) and Negative Learning (NL). To verify the effectiveness of the proposed scheme, we designed a portable ECG patch -- iRealCare -- and applied the algorithm on a real-life dataset. Our experimental results show that we can achieve an accuracy of 91.0 %, which is 6.2 % higher than a normal training process with ResNet. There are 65 patients in our dataset and we randomly picked 2 patients to perform validation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge