Shamik Bose
AgenTRIM: Tool Risk Mitigation for Agentic AI
Jan 18, 2026Abstract:AI agents are autonomous systems that combine LLMs with external tools to solve complex tasks. While such tools extend capability, improper tool permissions introduce security risks such as indirect prompt injection and tool misuse. We characterize these failures as unbalanced tool-driven agency. Agents may retain unnecessary permissions (excessive agency) or fail to invoke required tools (insufficient agency), amplifying the attack surface and reducing performance. We introduce AgenTRIM, a framework for detecting and mitigating tool-driven agency risks without altering an agent's internal reasoning. AgenTRIM addresses these risks through complementary offline and online phases. Offline, AgenTRIM reconstructs and verifies the agent's tool interface from code and execution traces. At runtime, it enforces per-step least-privilege tool access through adaptive filtering and status-aware validation of tool calls. Evaluating on the AgentDojo benchmark, AgenTRIM substantially reduces attack success while maintaining high task performance. Additional experiments show robustness to description-based attacks and effective enforcement of explicit safety policies. Together, these results demonstrate that AgenTRIM provides a practical, capability-preserving approach to safer tool use in LLM-based agents.
LumiMAS: A Comprehensive Framework for Real-Time Monitoring and Enhanced Observability in Multi-Agent Systems
Aug 17, 2025Abstract:The incorporation of large language models in multi-agent systems (MASs) has the potential to significantly improve our ability to autonomously solve complex problems. However, such systems introduce unique challenges in monitoring, interpreting, and detecting system failures. Most existing MAS observability frameworks focus on analyzing each individual agent separately, overlooking failures associated with the entire MAS. To bridge this gap, we propose LumiMAS, a novel MAS observability framework that incorporates advanced analytics and monitoring techniques. The proposed framework consists of three key components: a monitoring and logging layer, anomaly detection layer, and anomaly explanation layer. LumiMAS's first layer monitors MAS executions, creating detailed logs of the agents' activity. These logs serve as input to the anomaly detection layer, which detects anomalies across the MAS workflow in real time. Then, the anomaly explanation layer performs classification and root cause analysis (RCA) of the detected anomalies. LumiMAS was evaluated on seven different MAS applications, implemented using two popular MAS platforms, and a diverse set of possible failures. The applications include two novel failure-tailored applications that illustrate the effects of a hallucination or bias on the MAS. The evaluation results demonstrate LumiMAS's effectiveness in failure detection, classification, and RCA.
MAPS: A Multilingual Benchmark for Global Agent Performance and Security
May 21, 2025Abstract:Agentic AI systems, which build on Large Language Models (LLMs) and interact with tools and memory, have rapidly advanced in capability and scope. Yet, since LLMs have been shown to struggle in multilingual settings, typically resulting in lower performance and reduced safety, agentic systems risk inheriting these limitations. This raises concerns about the global accessibility of such systems, as users interacting in languages other than English may encounter unreliable or security-critical agent behavior. Despite growing interest in evaluating agentic AI, existing benchmarks focus exclusively on English, leaving multilingual settings unexplored. To address this gap, we propose MAPS, a multilingual benchmark suite designed to evaluate agentic AI systems across diverse languages and tasks. MAPS builds on four widely used agentic benchmarks - GAIA (real-world tasks), SWE-bench (code generation), MATH (mathematical reasoning), and the Agent Security Benchmark (security). We translate each dataset into ten diverse languages, resulting in 805 unique tasks and 8,855 total language-specific instances. Our benchmark suite enables a systematic analysis of how multilingual contexts affect agent performance and robustness. Empirically, we observe consistent degradation in both performance and security when transitioning from English to other languages, with severity varying by task and correlating with the amount of translated input. Building on these findings, we provide actionable recommendations to guide agentic AI systems development and assessment under multilingual settings. This work establishes a standardized evaluation framework, encouraging future research towards equitable, reliable, and globally accessible agentic AI. MAPS benchmark suite is publicly available at https://huggingface.co/datasets/Fujitsu-FRE/MAPS
The BigScience ROOTS Corpus: A 1.6TB Composite Multilingual Dataset
Mar 07, 2023



Abstract:As language models grow ever larger, the need for large-scale high-quality text datasets has never been more pressing, especially in multilingual settings. The BigScience workshop, a 1-year international and multidisciplinary initiative, was formed with the goal of researching and training large language models as a values-driven undertaking, putting issues of ethics, harm, and governance in the foreground. This paper documents the data creation and curation efforts undertaken by BigScience to assemble the Responsible Open-science Open-collaboration Text Sources (ROOTS) corpus, a 1.6TB dataset spanning 59 languages that was used to train the 176-billion-parameter BigScience Large Open-science Open-access Multilingual (BLOOM) language model. We further release a large initial subset of the corpus and analyses thereof, and hope to empower large-scale monolingual and multilingual modeling projects with both the data and the processing tools, as well as stimulate research around this large multilingual corpus.
SantaCoder: don't reach for the stars!
Jan 09, 2023



Abstract:The BigCode project is an open-scientific collaboration working on the responsible development of large language models for code. This tech report describes the progress of the collaboration until December 2022, outlining the current state of the Personally Identifiable Information (PII) redaction pipeline, the experiments conducted to de-risk the model architecture, and the experiments investigating better preprocessing methods for the training data. We train 1.1B parameter models on the Java, JavaScript, and Python subsets of The Stack and evaluate them on the MultiPL-E text-to-code benchmark. We find that more aggressive filtering of near-duplicates can further boost performance and, surprisingly, that selecting files from repositories with 5+ GitHub stars deteriorates performance significantly. Our best model outperforms previous open-source multilingual code generation models (InCoder-6.7B and CodeGen-Multi-2.7B) in both left-to-right generation and infilling on the Java, JavaScript, and Python portions of MultiPL-E, despite being a substantially smaller model. All models are released under an OpenRAIL license at https://hf.co/bigcode.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022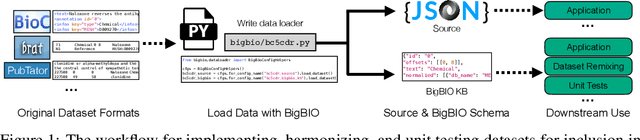
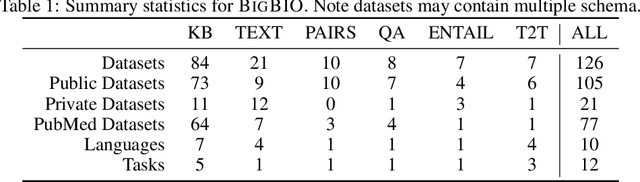
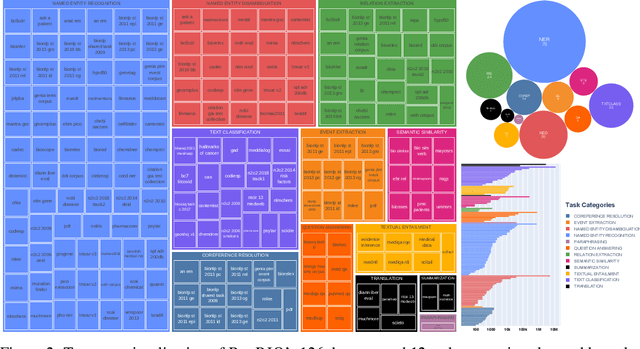
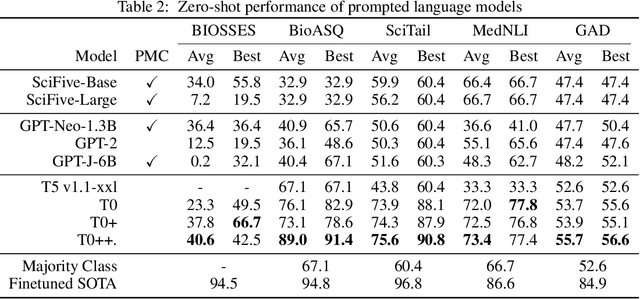
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge