Jan Deprest
DEEPBEAS3D: Deep Learning and B-Spline Explicit Active Surfaces
Sep 05, 2023Abstract:Deep learning-based automatic segmentation methods have become state-of-the-art. However, they are often not robust enough for direct clinical application, as domain shifts between training and testing data affect their performance. Failure in automatic segmentation can cause sub-optimal results that require correction. To address these problems, we propose a novel 3D extension of an interactive segmentation framework that represents a segmentation from a convolutional neural network (CNN) as a B-spline explicit active surface (BEAS). BEAS ensures segmentations are smooth in 3D space, increasing anatomical plausibility, while allowing the user to precisely edit the 3D surface. We apply this framework to the task of 3D segmentation of the anal sphincter complex (AS) from transperineal ultrasound (TPUS) images, and compare it to the clinical tool used in the pelvic floor disorder clinic (4D View VOCAL, GE Healthcare; Zipf, Austria). Experimental results show that: 1) the proposed framework gives the user explicit control of the surface contour; 2) the perceived workload calculated via the NASA-TLX index was reduced by 30% compared to VOCAL; and 3) it required 7 0% (170 seconds) less user time than VOCAL (p< 0.00001)
Adaptive Multi-scale Online Likelihood Network for AI-assisted Interactive Segmentation
Mar 23, 2023Abstract:Existing interactive segmentation methods leverage automatic segmentation and user interactions for label refinement, significantly reducing the annotation workload compared to manual annotation. However, these methods lack quick adaptability to ambiguous and noisy data, which is a challenge in CT volumes containing lung lesions from COVID-19 patients. In this work, we propose an adaptive multi-scale online likelihood network (MONet) that adaptively learns in a data-efficient online setting from both an initial automatic segmentation and user interactions providing corrections. We achieve adaptive learning by proposing an adaptive loss that extends the influence of user-provided interaction to neighboring regions with similar features. In addition, we propose a data-efficient probability-guided pruning method that discards uncertain and redundant labels in the initial segmentation to enable efficient online training and inference. Our proposed method was evaluated by an expert in a blinded comparative study on COVID-19 lung lesion annotation task in CT. Our approach achieved 5.86% higher Dice score with 24.67% less perceived NASA-TLX workload score than the state-of-the-art. Source code is available at: https://github.com/masadcv/MONet-MONAILabel
Retrieval of surgical phase transitions using reinforcement learning
Aug 01, 2022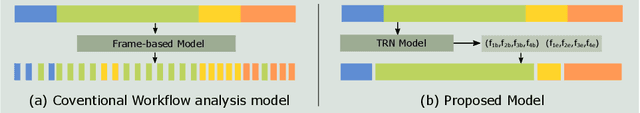
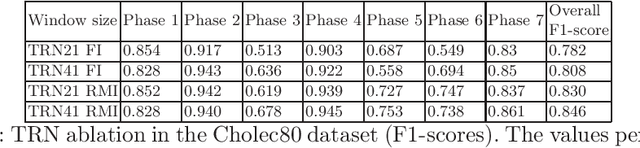
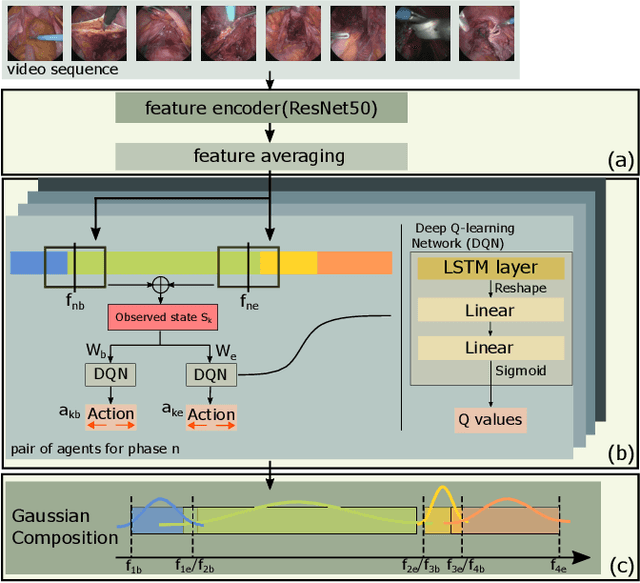
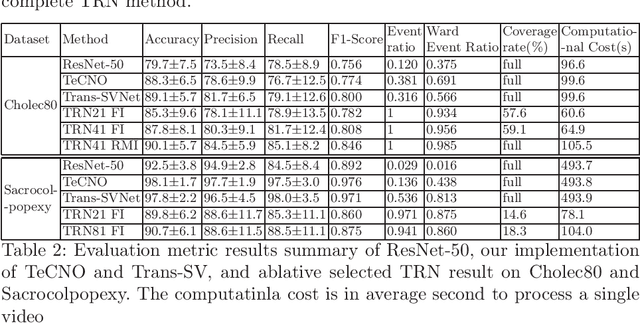
Abstract:In minimally invasive surgery, surgical workflow segmentation from video analysis is a well studied topic. The conventional approach defines it as a multi-class classification problem, where individual video frames are attributed a surgical phase label. We introduce a novel reinforcement learning formulation for offline phase transition retrieval. Instead of attempting to classify every video frame, we identify the timestamp of each phase transition. By construction, our model does not produce spurious and noisy phase transitions, but contiguous phase blocks. We investigate two different configurations of this model. The first does not require processing all frames in a video (only <60% and <20% of frames in 2 different applications), while producing results slightly under the state-of-the-art accuracy. The second configuration processes all video frames, and outperforms the state-of-the art at a comparable computational cost. We compare our method against the recent top-performing frame-based approaches TeCNO and Trans-SVNet on the public dataset Cholec80 and also on an in-house dataset of laparoscopic sacrocolpopexy. We perform both a frame-based (accuracy, precision, recall and F1-score) and an event-based (event ratio) evaluation of our algorithms.
Learning-Based Keypoint Registration for Fetoscopic Mosaicking
Jul 26, 2022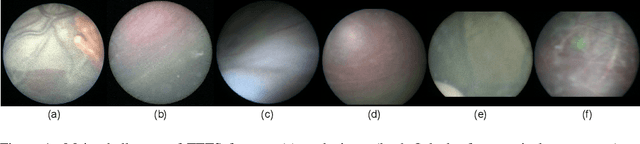
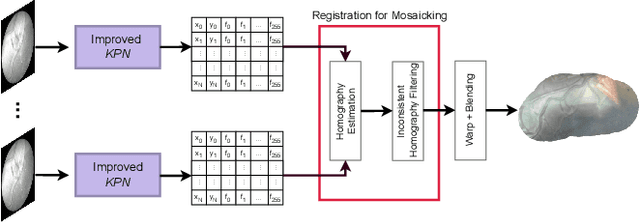

Abstract:In Twin-to-Twin Transfusion Syndrome (TTTS), abnormal vascular anastomoses in the monochorionic placenta can produce uneven blood flow between the two fetuses. In the current practice, TTTS is treated surgically by closing abnormal anastomoses using laser ablation. This surgery is minimally invasive and relies on fetoscopy. Limited field of view makes anastomosis identification a challenging task for the surgeon. To tackle this challenge, we propose a learning-based framework for in-vivo fetoscopy frame registration for field-of-view expansion. The novelties of this framework relies on a learning-based keypoint proposal network and an encoding strategy to filter (i) irrelevant keypoints based on fetoscopic image segmentation and (ii) inconsistent homographies. We validate of our framework on a dataset of 6 intraoperative sequences from 6 TTTS surgeries from 6 different women against the most recent state of the art algorithm, which relies on the segmentation of placenta vessels. The proposed framework achieves higher performance compared to the state of the art, paving the way for robust mosaicking to provide surgeons with context awareness during TTTS surgery.
FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy
Jun 30, 2022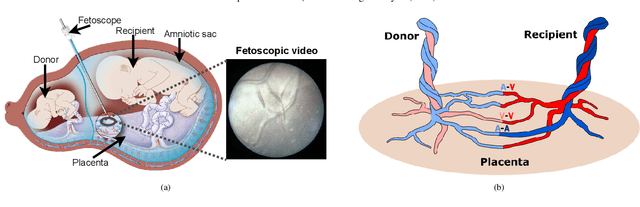
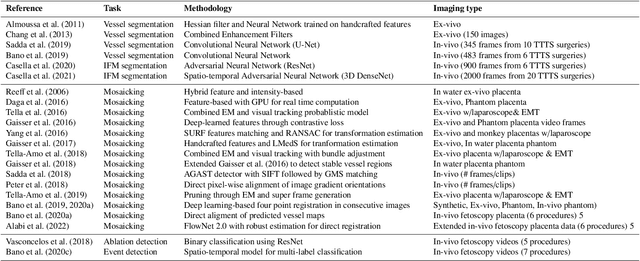
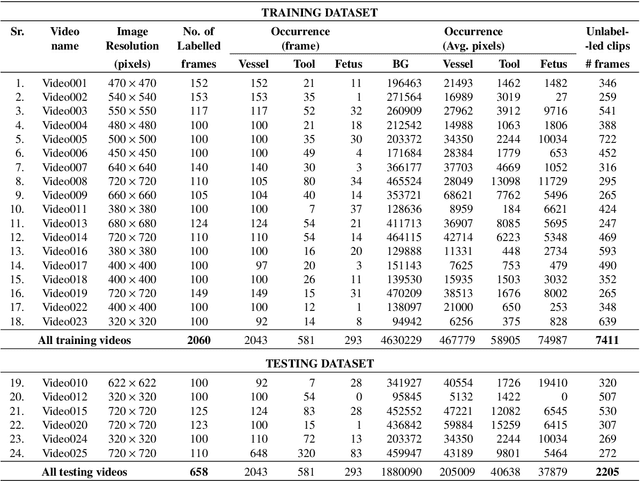
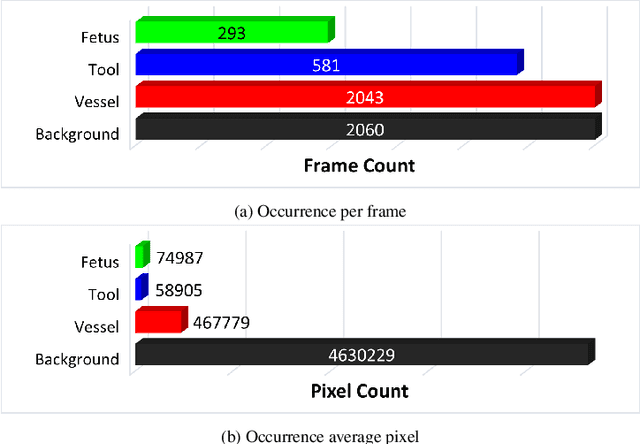
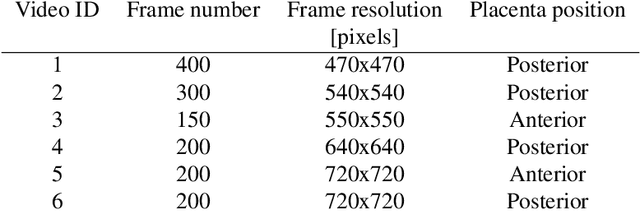
Abstract:Fetoscopy laser photocoagulation is a widely adopted procedure for treating Twin-to-Twin Transfusion Syndrome (TTTS). The procedure involves photocoagulation pathological anastomoses to regulate blood exchange among twins. The procedure is particularly challenging due to the limited field of view, poor manoeuvrability of the fetoscope, poor visibility, and variability in illumination. These challenges may lead to increased surgery time and incomplete ablation. Computer-assisted intervention (CAI) can provide surgeons with decision support and context awareness by identifying key structures in the scene and expanding the fetoscopic field of view through video mosaicking. Research in this domain has been hampered by the lack of high-quality data to design, develop and test CAI algorithms. Through the Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge, which was organized as part of the MICCAI2021 Endoscopic Vision challenge, we released the first largescale multicentre TTTS dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms. For this challenge, we released a dataset of 2060 images, pixel-annotated for vessels, tool, fetus and background classes, from 18 in-vivo TTTS fetoscopy procedures and 18 short video clips. Seven teams participated in this challenge and their model performance was assessed on an unseen test dataset of 658 pixel-annotated images from 6 fetoscopic procedures and 6 short clips. The challenge provided an opportunity for creating generalized solutions for fetoscopic scene understanding and mosaicking. In this paper, we present the findings of the FetReg2021 challenge alongside reporting a detailed literature review for CAI in TTTS fetoscopy. Through this challenge, its analysis and the release of multi-centre fetoscopic data, we provide a benchmark for future research in this field.
A Dempster-Shafer approach to trustworthy AI with application to fetal brain MRI segmentation
Apr 05, 2022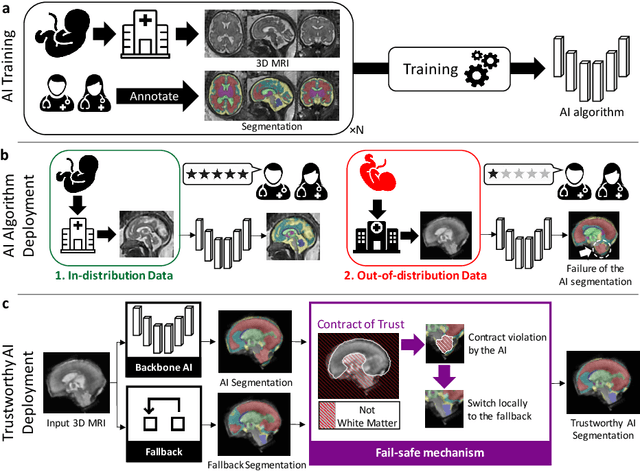
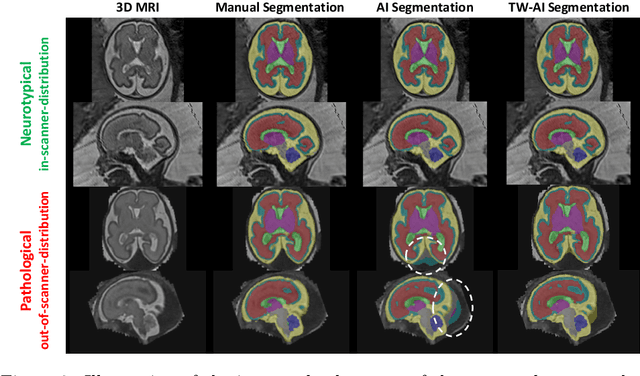
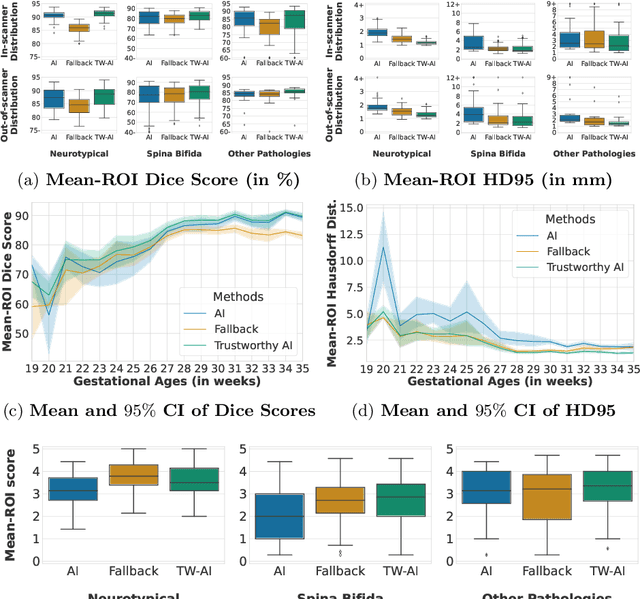
Abstract:Deep learning models for medical image segmentation can fail unexpectedly and spectacularly for pathological cases and for images acquired at different centers than those used for training, with labeling errors that violate expert knowledge about the anatomy and the intensity distribution of the regions to be segmented. Such errors undermine the trustworthiness of deep learning models developed for medical image segmentation. Mechanisms with a fallback method for detecting and correcting such failures are essential for safely translating this technology into clinics and are likely to be a requirement of future regulations on artificial intelligence (AI). Here, we propose a principled trustworthy AI theoretical framework and a practical system that can augment any backbone AI system using a fallback method and a fail-safe mechanism based on Dempster-Shafer theory. Our approach relies on an actionable definition of trustworthy AI. Our method automatically discards the voxel-level labeling predicted by the backbone AI that are likely to violate expert knowledge and relies on a fallback atlas-based segmentation method for those voxels. We demonstrate the effectiveness of the proposed trustworthy AI approach on the largest reported annotated dataset of fetal T2w MRI consisting of 540 manually annotated fetal brain 3D MRIs with neurotypical or abnormal brain development and acquired from 13 sources of data across 6 countries. We show that our trustworthy AI method improves the robustness of a state-of-the-art backbone AI for fetal brain MRI segmentation on MRIs acquired across various centers and for fetuses with various brain abnormalities.
Partial supervision for the FeTA challenge 2021
Nov 03, 2021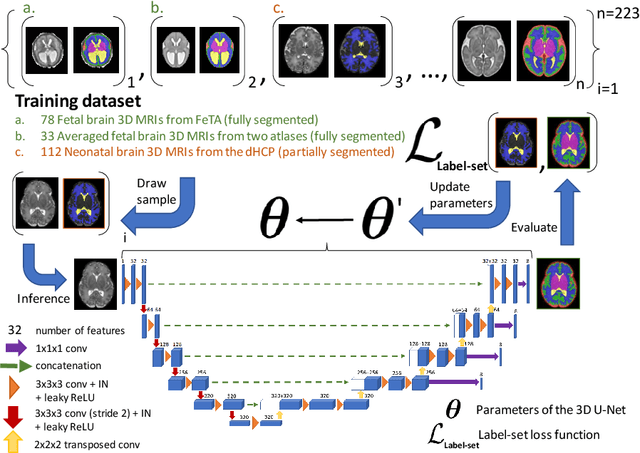
Abstract:This paper describes our method for our participation in the FeTA challenge2021 (team name: TRABIT). The performance of convolutional neural networks for medical image segmentation is thought to correlate positively with the number of training data. The FeTA challenge does not restrict participants to using only the provided training data but also allows for using other publicly available sources. Yet, open access fetal brain data remains limited. An advantageous strategy could thus be to expand the training data to cover broader perinatal brain imaging sources. Perinatal brain MRIs, other than the FeTA challenge data, that are currently publicly available, span normal and pathological fetal atlases as well as neonatal scans. However, perinatal brain MRIs segmented in different datasets typically come with different annotation protocols. This makes it challenging to combine those datasets to train a deep neural network. We recently proposed a family of loss functions, the label-set loss functions, for partially supervised learning. Label-set loss functions allow to train deep neural networks with partially segmented images, i.e. segmentations in which some classes may be grouped into super-classes. We propose to use label-set loss functions to improve the segmentation performance of a state-of-the-art deep learning pipeline for multi-class fetal brain segmentation by merging several publicly available datasets. To promote generalisability, our approach does not introduce any additional hyper-parameters tuning.
Interactive Segmentation via Deep Learning and B-Spline Explicit Active Surfaces
Oct 25, 2021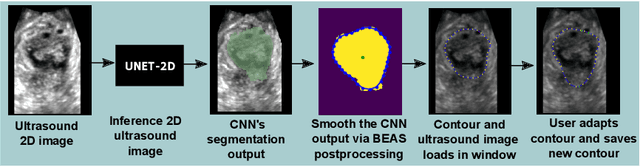
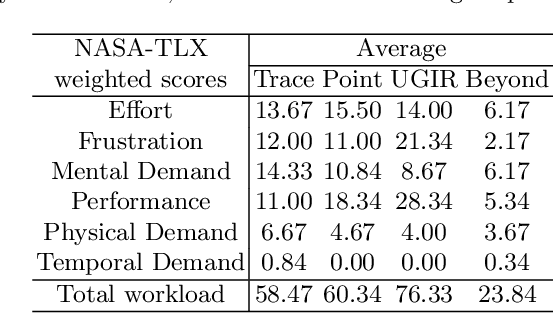
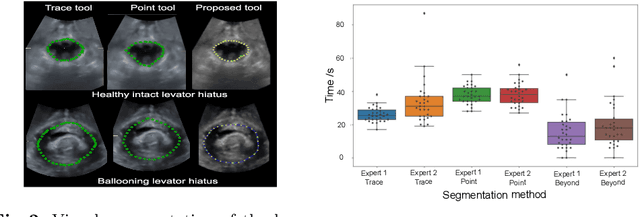
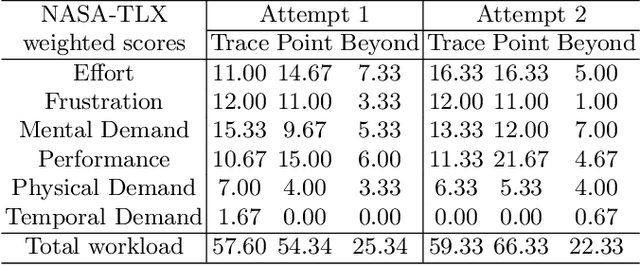
Abstract:Automatic medical image segmentation via convolutional neural networks (CNNs) has shown promising results. However, they may not always be robust enough for clinical use. Sub-optimal segmentation would require clinician's to manually delineate the target object, causing frustration. To address this problem, a novel interactive CNN-based segmentation framework is proposed in this work. The aim is to represent the CNN segmentation contour as B-splines by utilising B-spline explicit active surfaces (BEAS). The interactive element of the framework allows the user to precisely edit the contour in real-time, and by utilising BEAS it ensures the final contour is smooth and anatomically plausible. This framework was applied to the task of 2D segmentation of the levator hiatus from 2D ultrasound (US) images, and compared to the current clinical tools used in pelvic floor disorder clinic (4DView, GE Healthcare; Zipf, Austria). Experimental results show that: 1) the proposed framework is more robust than current state-of-the-art CNNs; 2) the perceived workload calculated via the NASA-TLX index was reduced more than half for the proposed approach in comparison to current clinical tools; and 3) the proposed tool requires at least 13 seconds less user time than the clinical tools, which was significant (p=0.001).
* 11 pages, 3 figures, 2 tables
Distributionally Robust Segmentation of Abnormal Fetal Brain 3D MRI
Aug 09, 2021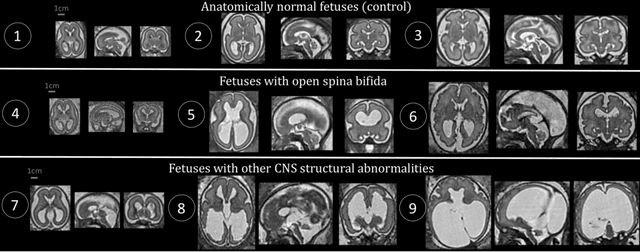
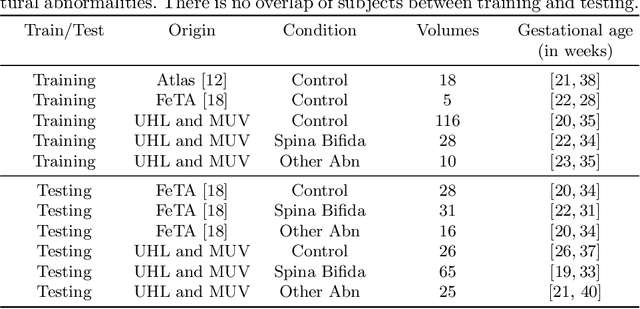
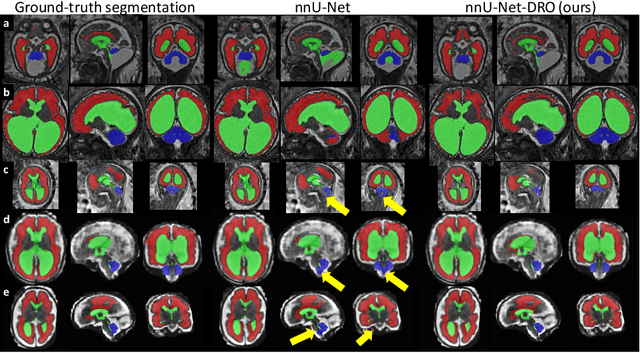
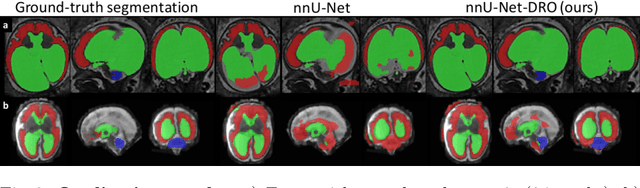
Abstract:The performance of deep neural networks typically increases with the number of training images. However, not all images have the same importance towards improved performance and robustness. In fetal brain MRI, abnormalities exacerbate the variability of the developing brain anatomy compared to non-pathological cases. A small number of abnormal cases, as is typically available in clinical datasets used for training, are unlikely to fairly represent the rich variability of abnormal developing brains. This leads machine learning systems trained by maximizing the average performance to be biased toward non-pathological cases. This problem was recently referred to as hidden stratification. To be suited for clinical use, automatic segmentation methods need to reliably achieve high-quality segmentation outcomes also for pathological cases. In this paper, we show that the state-of-the-art deep learning pipeline nnU-Net has difficulties to generalize to unseen abnormal cases. To mitigate this problem, we propose to train a deep neural network to minimize a percentile of the distribution of per-volume loss over the dataset. We show that this can be achieved by using Distributionally Robust Optimization (DRO). DRO automatically reweights the training samples with lower performance, encouraging nnU-Net to perform more consistently on all cases. We validated our approach using a dataset of 368 fetal brain T2w MRIs, including 124 MRIs of open spina bifida cases and 51 MRIs of cases with other severe abnormalities of brain development.
Label-set Loss Functions for Partial Supervision: Application to Fetal Brain 3D MRI Parcellation
Jul 09, 2021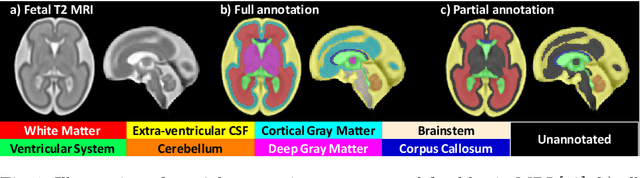
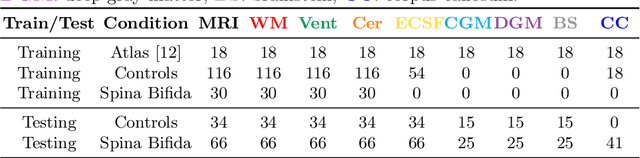
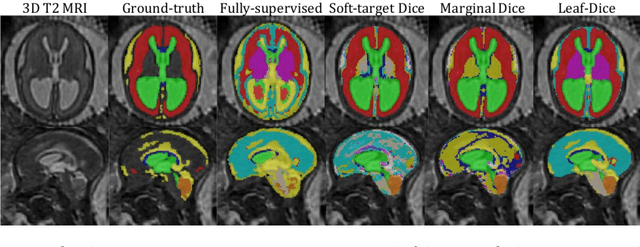
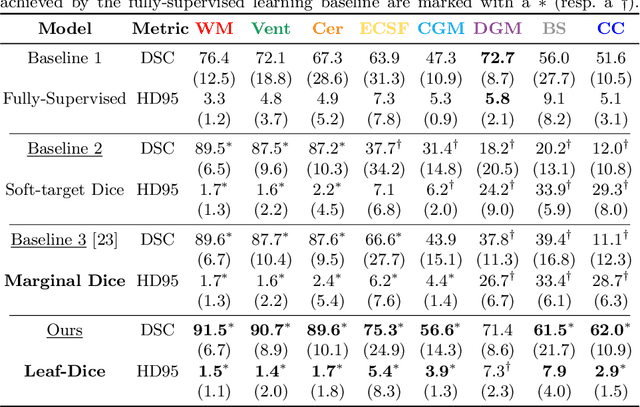
Abstract:Deep neural networks have increased the accuracy of automatic segmentation, however, their accuracy depends on the availability of a large number of fully segmented images. Methods to train deep neural networks using images for which some, but not all, regions of interest are segmented are necessary to make better use of partially annotated datasets. In this paper, we propose the first axiomatic definition of label-set loss functions that are the loss functions that can handle partially segmented images. We prove that there is one and only one method to convert a classical loss function for fully segmented images into a proper label-set loss function. Our theory also allows us to define the leaf-Dice loss, a label-set generalization of the Dice loss particularly suited for partial supervision with only missing labels. Using the leaf-Dice loss, we set a new state of the art in partially supervised learning for fetal brain 3D MRI segmentation. We achieve a deep neural network able to segment white matter, ventricles, cerebellum, extra-ventricular CSF, cortical gray matter, deep gray matter, brainstem, and corpus callosum based on fetal brain 3D MRI of anatomically normal fetuses or with open spina bifida. Our implementation of the proposed label-set loss functions is available at https://github.com/LucasFidon/label-set-loss-functions
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge