Pengfei Guo
MoViT: Memorizing Vision Transformers for Medical Image Analysis
Apr 04, 2023



Abstract:The synergy of long-range dependencies from transformers and local representations of image content from convolutional neural networks (CNNs) has led to advanced architectures and increased performance for various medical image analysis tasks due to their complementary benefits. However, compared with CNNs, transformers require considerably more training data, due to a larger number of parameters and an absence of inductive bias. The need for increasingly large datasets continues to be problematic, particularly in the context of medical imaging, where both annotation efforts and data protection result in limited data availability. In this work, inspired by the human decision-making process of correlating new ``evidence'' with previously memorized ``experience'', we propose a Memorizing Vision Transformer (MoViT) to alleviate the need for large-scale datasets to successfully train and deploy transformer-based architectures. MoViT leverages an external memory structure to cache history attention snapshots during the training stage. To prevent overfitting, we incorporate an innovative memory update scheme, attention temporal moving average, to update the stored external memories with the historical moving average. For inference speedup, we design a prototypical attention learning method to distill the external memory into smaller representative subsets. We evaluate our method on a public histology image dataset and an in-house MRI dataset, demonstrating that MoViT applied to varied medical image analysis tasks, can outperform vanilla transformer models across varied data regimes, especially in cases where only a small amount of annotated data is available. More importantly, MoViT can reach a competitive performance of ViT with only 3.0% of the training data.
Deep-learning-enabled Brain Hemodynamic Mapping Using Resting-state fMRI
Apr 25, 2022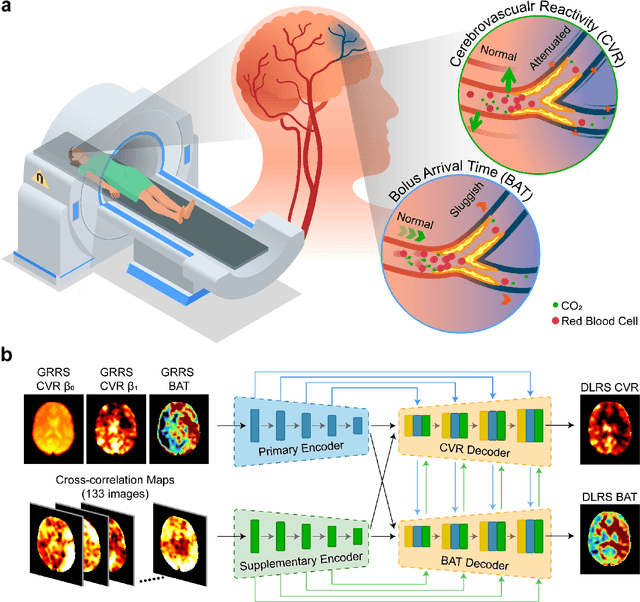
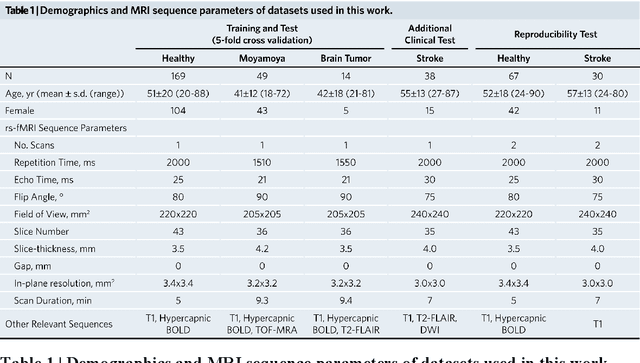
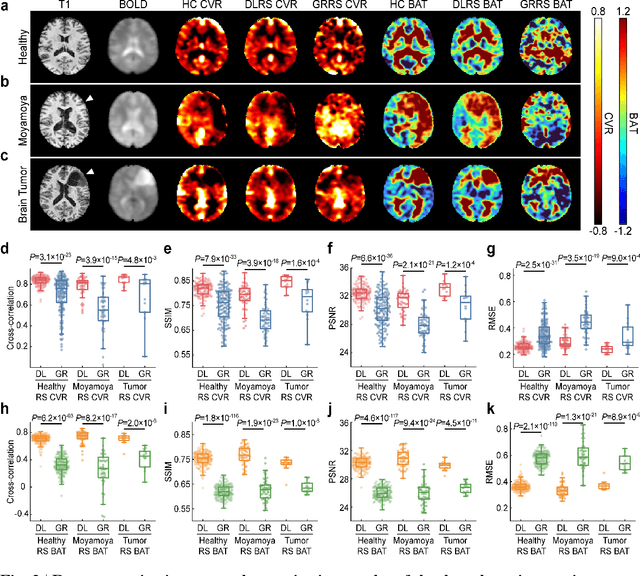
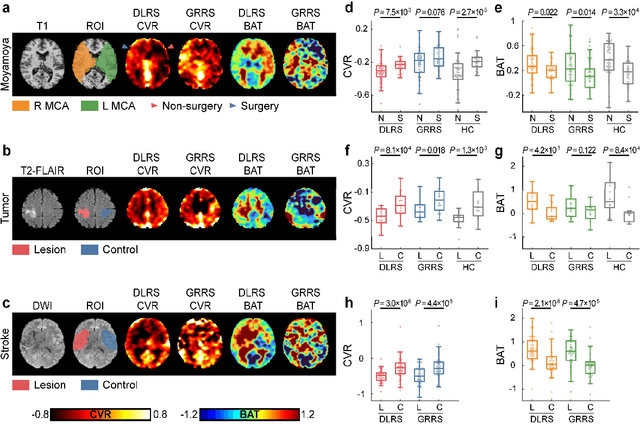
Abstract:Cerebrovascular disease is a leading cause of death globally. Prevention and early intervention are known to be the most effective forms of its management. Non-invasive imaging methods hold great promises for early stratification, but at present lack the sensitivity for personalized prognosis. Resting-state functional magnetic resonance imaging (rs-fMRI), a powerful tool previously used for mapping neural activity, is available in most hospitals. Here we show that rs-fMRI can be used to map cerebral hemodynamic function and delineate impairment. By exploiting time variations in breathing pattern during rs-fMRI, deep learning enables reproducible mapping of cerebrovascular reactivity (CVR) and bolus arrive time (BAT) of the human brain using resting-state CO2 fluctuations as a natural 'contrast media'. The deep-learning network was trained with CVR and BAT maps obtained with a reference method of CO2-inhalation MRI, which included data from young and older healthy subjects and patients with Moyamoya disease and brain tumors. We demonstrate the performance of deep-learning cerebrovascular mapping in the detection of vascular abnormalities, evaluation of revascularization effects, and vascular alterations in normal aging. In addition, cerebrovascular maps obtained with the proposed method exhibited excellent reproducibility in both healthy volunteers and stroke patients. Deep-learning resting-state vascular imaging has the potential to become a useful tool in clinical cerebrovascular imaging.
Escaping Data Scarcity for High-Resolution Heterogeneous Face Hallucination
Mar 30, 2022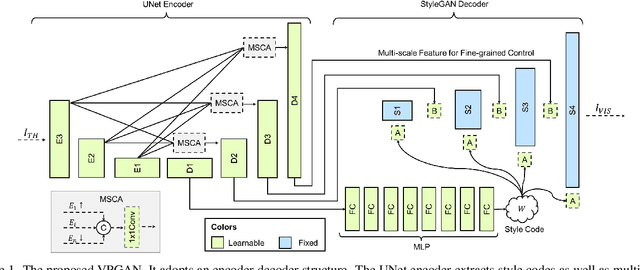
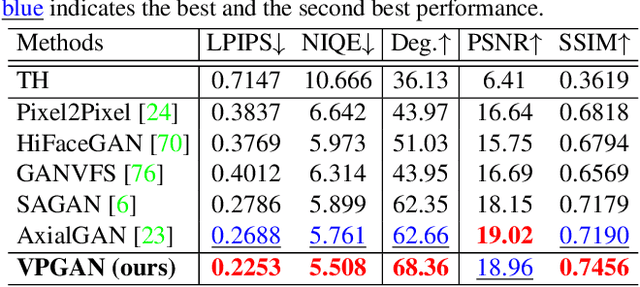
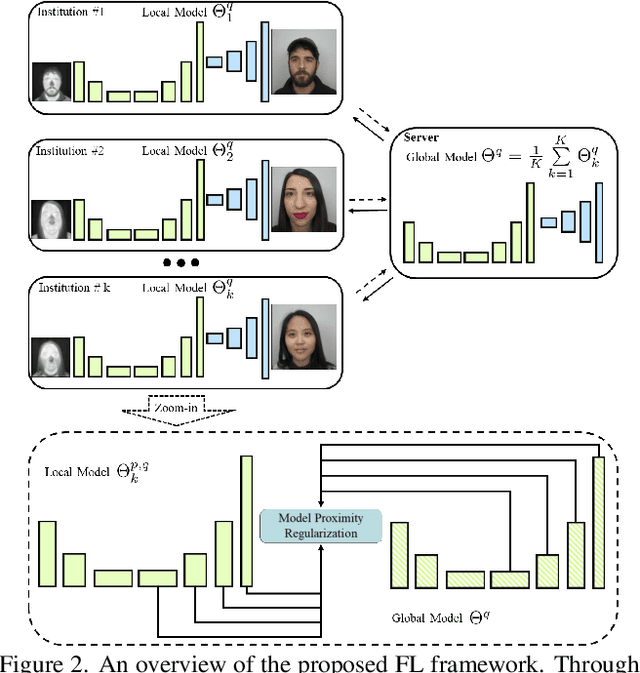
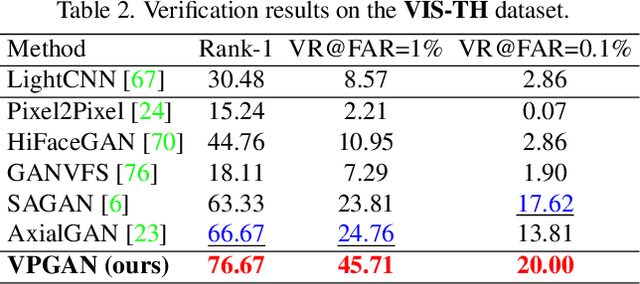
Abstract:In Heterogeneous Face Recognition (HFR), the objective is to match faces across two different domains such as visible and thermal. Large domain discrepancy makes HFR a difficult problem. Recent methods attempting to fill the gap via synthesis have achieved promising results, but their performance is still limited by the scarcity of paired training data. In practice, large-scale heterogeneous face data are often inaccessible due to the high cost of acquisition and annotation process as well as privacy regulations. In this paper, we propose a new face hallucination paradigm for HFR, which not only enables data-efficient synthesis but also allows to scale up model training without breaking any privacy policy. Unlike existing methods that learn face synthesis entirely from scratch, our approach is particularly designed to take advantage of rich and diverse facial priors from visible domain for more faithful hallucination. On the other hand, large-scale training is enabled by introducing a new federated learning scheme to allow institution-wise collaborations while avoiding explicit data sharing. Extensive experiments demonstrate the advantages of our approach in tackling HFR under current data limitations. In a unified framework, our method yields the state-of-the-art hallucination results on multiple HFR datasets.
Closing the Generalization Gap of Cross-silo Federated Medical Image Segmentation
Mar 18, 2022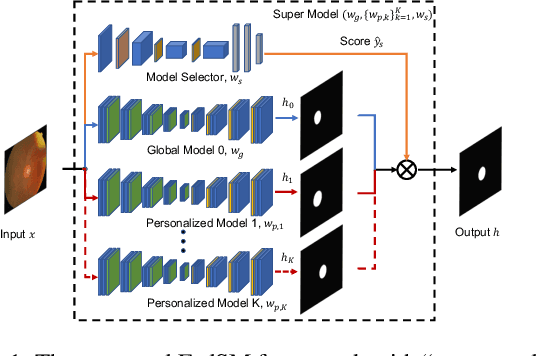
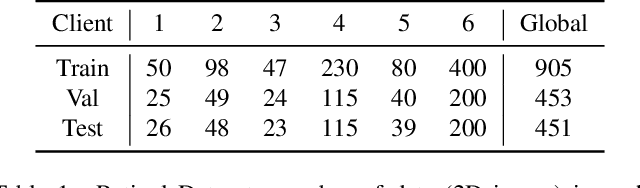
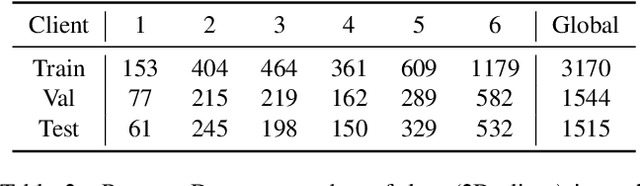
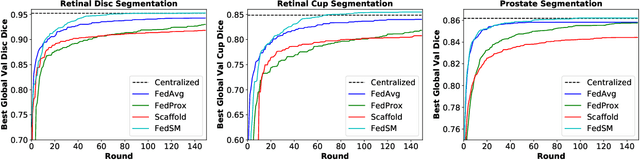
Abstract:Cross-silo federated learning (FL) has attracted much attention in medical imaging analysis with deep learning in recent years as it can resolve the critical issues of insufficient data, data privacy, and training efficiency. However, there can be a generalization gap between the model trained from FL and the one from centralized training. This important issue comes from the non-iid data distribution of the local data in the participating clients and is well-known as client drift. In this work, we propose a novel training framework FedSM to avoid the client drift issue and successfully close the generalization gap compared with the centralized training for medical image segmentation tasks for the first time. We also propose a novel personalized FL objective formulation and a new method SoftPull to solve it in our proposed framework FedSM. We conduct rigorous theoretical analysis to guarantee its convergence for optimizing the non-convex smooth objective function. Real-world medical image segmentation experiments using deep FL validate the motivations and effectiveness of our proposed method.
Auto-FedRL: Federated Hyperparameter Optimization for Multi-institutional Medical Image Segmentation
Mar 12, 2022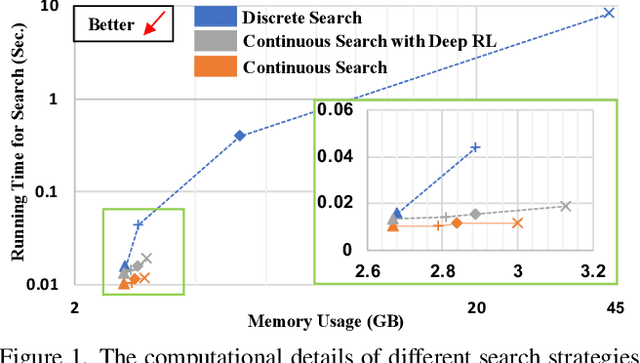
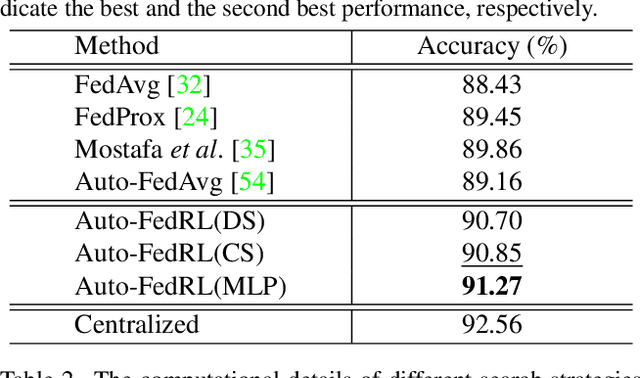
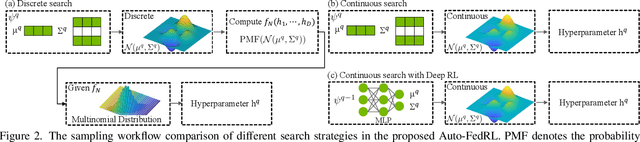

Abstract:Federated learning (FL) is a distributed machine learning technique that enables collaborative model training while avoiding explicit data sharing. The inherent privacy-preserving property of FL algorithms makes them especially attractive to the medical field. However, in case of heterogeneous client data distributions, standard FL methods are unstable and require intensive hyperparameter tuning to achieve optimal performance. Conventional hyperparameter optimization algorithms are impractical in real-world FL applications as they involve numerous training trials, which are often not affordable with limited compute budgets. In this work, we propose an efficient reinforcement learning~(RL)-based federated hyperparameter optimization algorithm, termed Auto-FedRL, in which an online RL agent can dynamically adjust hyperparameters of each client based on the current training progress. Extensive experiments are conducted to investigate different search strategies and RL agents. The effectiveness of the proposed method is validated on a heterogeneous data split of the CIFAR-10 dataset as well as two real-world medical image segmentation datasets for COVID-19 lesion segmentation in chest CT and pancreas segmentation in abdominal CT.
Towards performant and reliable undersampled MR reconstruction via diffusion model sampling
Mar 11, 2022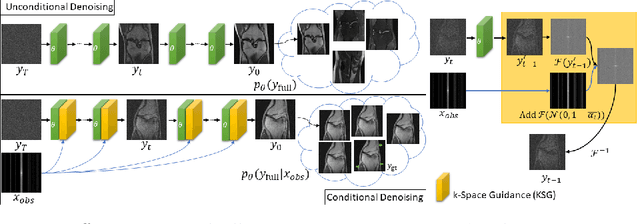
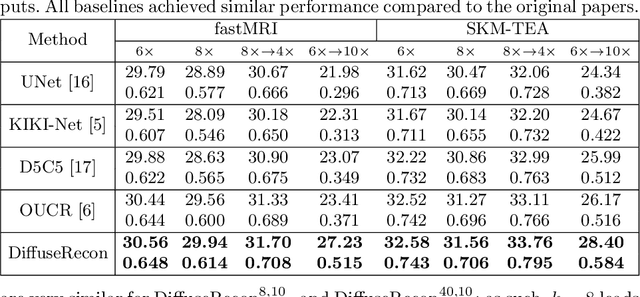
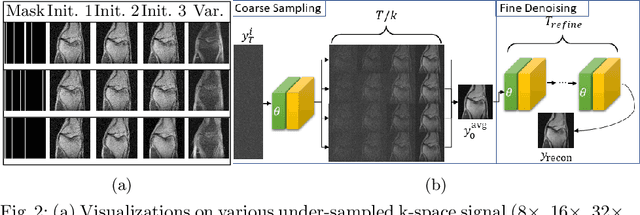
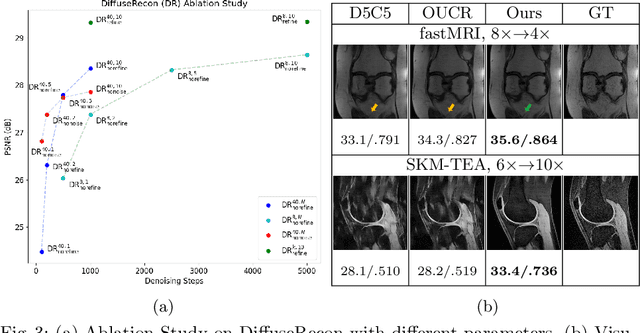
Abstract:Magnetic Resonance (MR) image reconstruction from under-sampled acquisition promises faster scanning time. To this end, current State-of-The-Art (SoTA) approaches leverage deep neural networks and supervised training to learn a recovery model. While these approaches achieve impressive performances, the learned model can be fragile on unseen degradation, e.g. when given a different acceleration factor. These methods are also generally deterministic and provide a single solution to an ill-posed problem; as such, it can be difficult for practitioners to understand the reliability of the reconstruction. We introduce DiffuseRecon, a novel diffusion model-based MR reconstruction method. DiffuseRecon guides the generation process based on the observed signals and a pre-trained diffusion model, and does not require additional training on specific acceleration factors. DiffuseRecon is stochastic in nature and generates results from a distribution of fully-sampled MR images; as such, it allows us to explicitly visualize different potential reconstruction solutions. Lastly, DiffuseRecon proposes an accelerated, coarse-to-fine Monte-Carlo sampling scheme to approximate the most likely reconstruction candidate. The proposed DiffuseRecon achieves SoTA performances reconstructing from raw acquisition signals in fastMRI and SKM-TEA. Code will be open-sourced at www.github.com/cpeng93/DiffuseRecon.
On-the-Fly Test-time Adaptation for Medical Image Segmentation
Mar 10, 2022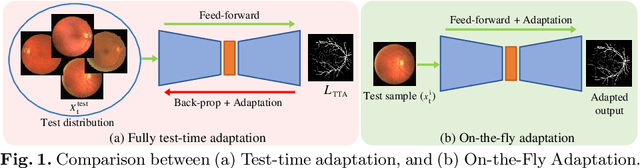

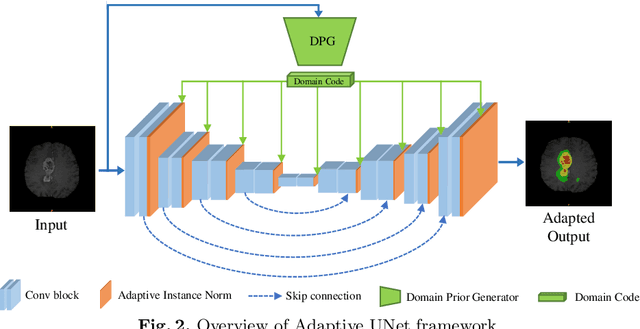
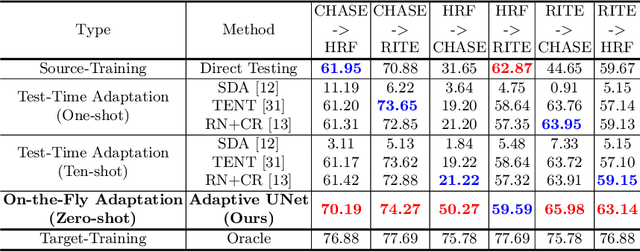
Abstract:One major problem in deep learning-based solutions for medical imaging is the drop in performance when a model is tested on a data distribution different from the one that it is trained on. Adapting the source model to target data distribution at test-time is an efficient solution for the data-shift problem. Previous methods solve this by adapting the model to target distribution by using techniques like entropy minimization or regularization. In these methods, the models are still updated by back-propagation using an unsupervised loss on complete test data distribution. In real-world clinical settings, it makes more sense to adapt a model to a new test image on-the-fly and avoid model update during inference due to privacy concerns and lack of computing resource at deployment. To this end, we propose a new setting - On-the-Fly Adaptation which is zero-shot and episodic (i.e., the model is adapted to a single image at a time and also does not perform any back-propagation during test-time). To achieve this, we propose a new framework called Adaptive UNet where each convolutional block is equipped with an adaptive batch normalization layer to adapt the features with respect to a domain code. The domain code is generated using a pre-trained encoder trained on a large corpus of medical images. During test-time, the model takes in just the new test image and generates a domain code to adapt the features of source model according to the test data. We validate the performance on both 2D and 3D data distribution shifts where we get a better performance compared to previous test-time adaptation methods. Code is available at https://github.com/jeya-maria-jose/On-The-Fly-Adaptation
ReconFormer: Accelerated MRI Reconstruction Using Recurrent Transformer
Jan 28, 2022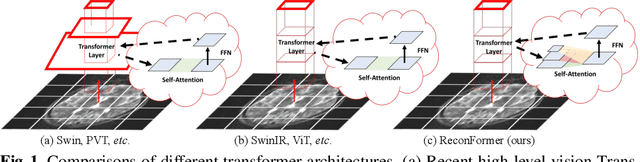
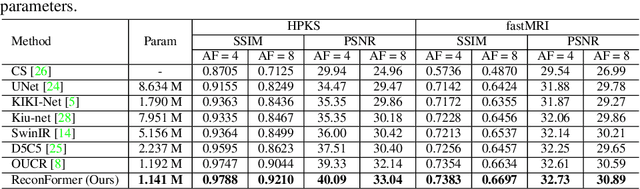
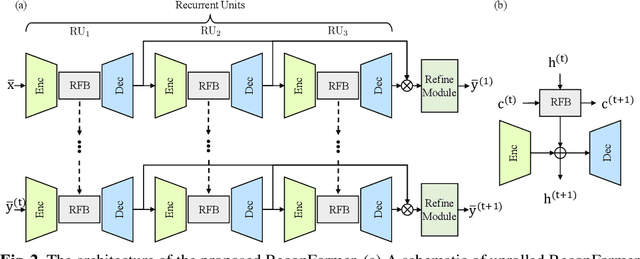
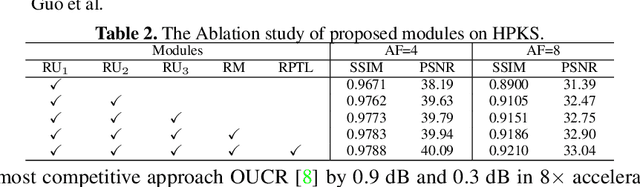
Abstract:Accelerating magnetic resonance image (MRI) reconstruction process is a challenging ill-posed inverse problem due to the excessive under-sampling operation in k-space. In this paper, we propose a recurrent transformer model, namely ReconFormer, for MRI reconstruction which can iteratively reconstruct high fertility magnetic resonance images from highly under-sampled k-space data. In particular, the proposed architecture is built upon Recurrent Pyramid Transformer Layers (RPTL), which jointly exploits intrinsic multi-scale information at every architecture unit as well as the dependencies of the deep feature correlation through recurrent states. Moreover, the proposed ReconFormer is lightweight since it employs the recurrent structure for its parameter efficiency. We validate the effectiveness of ReconFormer on multiple datasets with different magnetic resonance sequences and show that it achieves significant improvements over the state-of-the-art methods with better parameter efficiency. Implementation code will be available in https://github.com/guopengf/ReconFormer.
Reference-based Magnetic Resonance Image Reconstruction Using Texture Transforme
Nov 18, 2021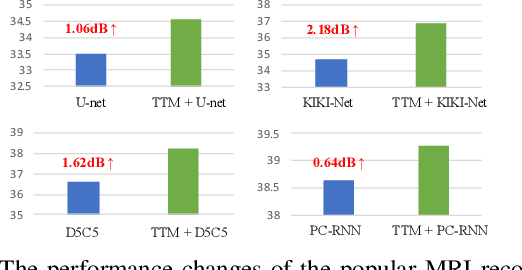
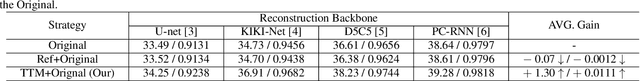
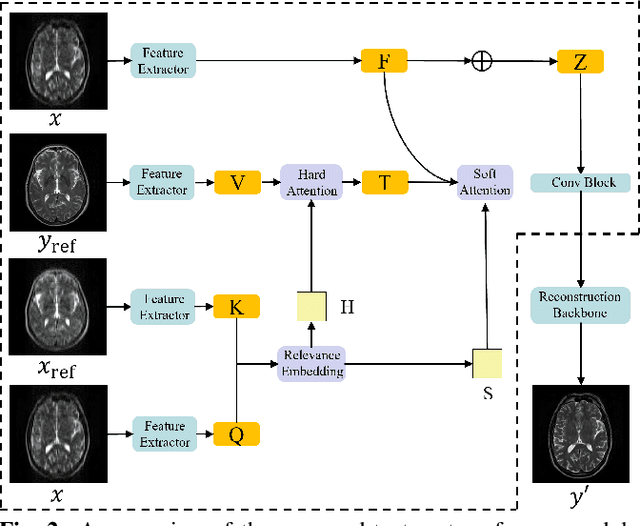
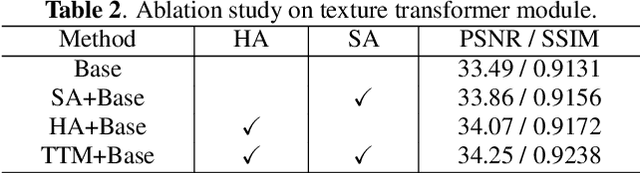
Abstract:Deep Learning (DL) based methods for magnetic resonance (MR) image reconstruction have been shown to produce superior performance in recent years. However, these methods either only leverage under-sampled data or require a paired fully-sampled auxiliary modality to perform multi-modal reconstruction. Consequently, existing approaches neglect to explore attention mechanisms that can transfer textures from reference fully-sampled data to under-sampled data within a single modality, which limits these approaches in challenging cases. In this paper, we propose a novel Texture Transformer Module (TTM) for accelerated MRI reconstruction, in which we formulate the under-sampled data and reference data as queries and keys in a transformer. The TTM facilitates joint feature learning across under-sampled and reference data, so the feature correspondences can be discovered by attention and accurate texture features can be leveraged during reconstruction. Notably, the proposed TTM can be stacked on prior MRI reconstruction approaches to further improve their performance. Extensive experiments show that TTM can significantly improve the performance of several popular DL-based MRI reconstruction methods.
Over-and-Under Complete Convolutional RNN for MRI Reconstruction
Jun 25, 2021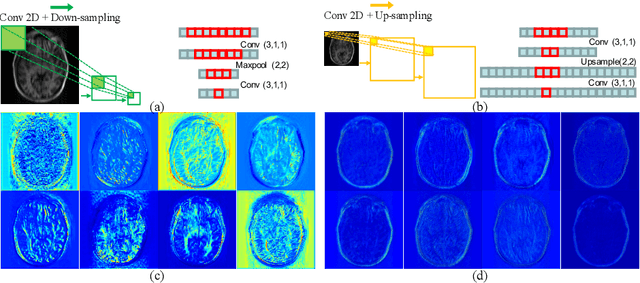
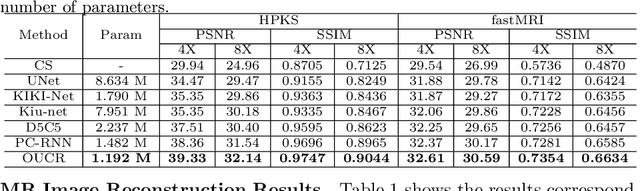
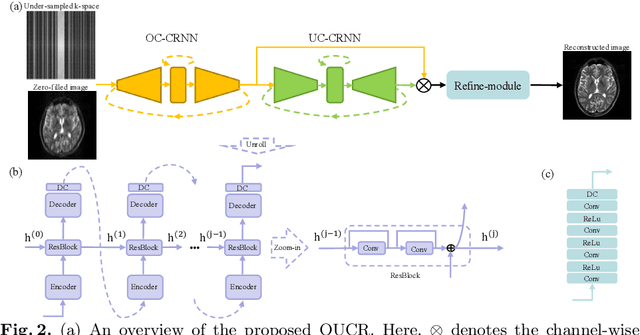
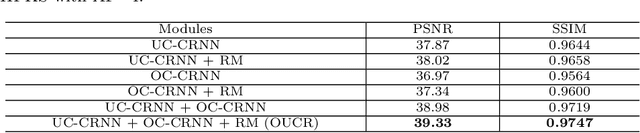
Abstract:Reconstructing magnetic resonance (MR) images from undersampled data is a challenging problem due to various artifacts introduced by the under-sampling operation. Recent deep learning-based methods for MR image reconstruction usually leverage a generic auto-encoder architecture which captures low-level features at the initial layers and high-level features at the deeper layers. Such networks focus much on global features which may not be optimal to reconstruct the fully-sampled image. In this paper, we propose an Over-and-Under Complete Convolutional Recurrent Neural Network (OUCR), which consists of an overcomplete and an undercomplete Convolutional Recurrent Neural Network(CRNN). The overcomplete branch gives special attention in learning local structures by restraining the receptive field of the network. Combining it with the undercomplete branch leads to a network which focuses more on low-level features without losing out on the global structures. Extensive experiments on two datasets demonstrate that the proposed method achieves significant improvements over the compressed sensing and popular deep learning-based methods with less number of trainable parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge