Ali Hatamizadeh
iGRPO: Self-Feedback-Driven LLM Reasoning
Feb 09, 2026Abstract:Large Language Models (LLMs) have shown promise in solving complex mathematical problems, yet they still fall short of producing accurate and consistent solutions. Reinforcement Learning (RL) is a framework for aligning these models with task-specific rewards, improving overall quality and reliability. Group Relative Policy Optimization (GRPO) is an efficient, value-function-free alternative to Proximal Policy Optimization (PPO) that leverages group-relative reward normalization. We introduce Iterative Group Relative Policy Optimization (iGRPO), a two-stage extension of GRPO that adds dynamic self-conditioning through model-generated drafts. In Stage 1, iGRPO samples multiple exploratory drafts and selects the highest-reward draft using the same scalar reward signal used for optimization. In Stage 2, it appends this best draft to the original prompt and applies a GRPO-style update on draft-conditioned refinements, training the policy to improve beyond its strongest prior attempt. Under matched rollout budgets, iGRPO consistently outperforms GRPO across base models (e.g., Nemotron-H-8B-Base-8K and DeepSeek-R1 Distilled), validating its effectiveness on diverse reasoning benchmarks. Moreover, applying iGRPO to OpenReasoning-Nemotron-7B trained on AceReason-Math achieves new state-of-the-art results of 85.62\% and 79.64\% on AIME24 and AIME25, respectively. Ablations further show that the refinement wrapper generalizes beyond GRPO variants, benefits from a generative judge, and alters learning dynamics by delaying entropy collapse. These results underscore the potential of iterative, self-feedback-based RL for advancing verifiable mathematical reasoning.
Gated Delta Networks: Improving Mamba2 with Delta Rule
Dec 09, 2024Abstract:Linear Transformers have gained attention as efficient alternatives to standard Transformers, but their performance in retrieval and long-context tasks has been limited. To address these limitations, recent work has explored two distinct mechanisms: gating for adaptive memory control and the delta update rule for precise memory modifications. We observe that these mechanisms are complementary: gating enables rapid memory erasure while the delta rule facilitates targeted updates. Building on this insight, we introduce the gated delta rule and develop a parallel training algorithm optimized for modern hardware. Our proposed architecture, Gated DeltaNet, consistently surpasses existing models like Mamba2 and DeltaNet across multiple benchmarks, including language modeling, common-sense reasoning, in-context retrieval, length extrapolation, and long-context understanding. We further enhance performance by developing hybrid architectures that combine Gated DeltaNet layers with sliding window attention or Mamba2 layers, achieving both improved training efficiency and superior task performance.
MambaVision: A Hybrid Mamba-Transformer Vision Backbone
Jul 10, 2024Abstract:We propose a novel hybrid Mamba-Transformer backbone, denoted as MambaVision, which is specifically tailored for vision applications. Our core contribution includes redesigning the Mamba formulation to enhance its capability for efficient modeling of visual features. In addition, we conduct a comprehensive ablation study on the feasibility of integrating Vision Transformers (ViT) with Mamba. Our results demonstrate that equipping the Mamba architecture with several self-attention blocks at the final layers greatly improves the modeling capacity to capture long-range spatial dependencies. Based on our findings, we introduce a family of MambaVision models with a hierarchical architecture to meet various design criteria. For Image classification on ImageNet-1K dataset, MambaVision model variants achieve a new State-of-the-Art (SOTA) performance in terms of Top-1 accuracy and image throughput. In downstream tasks such as object detection, instance segmentation and semantic segmentation on MS COCO and ADE20K datasets, MambaVision outperforms comparably-sized backbones and demonstrates more favorable performance. Code: https://github.com/NVlabs/MambaVision.
An Empirical Study of Mamba-based Language Models
Jun 12, 2024



Abstract:Selective state-space models (SSMs) like Mamba overcome some of the shortcomings of Transformers, such as quadratic computational complexity with sequence length and large inference-time memory requirements from the key-value cache. Moreover, recent studies have shown that SSMs can match or exceed the language modeling capabilities of Transformers, making them an attractive alternative. In a controlled setting (e.g., same data), however, studies so far have only presented small scale experiments comparing SSMs to Transformers. To understand the strengths and weaknesses of these architectures at larger scales, we present a direct comparison between 8B-parameter Mamba, Mamba-2, and Transformer models trained on the same datasets of up to 3.5T tokens. We also compare these models to a hybrid architecture consisting of 43% Mamba-2, 7% attention, and 50% MLP layers (Mamba-2-Hybrid). Using a diverse set of tasks, we answer the question of whether Mamba models can match Transformers at larger training budgets. Our results show that while pure SSMs match or exceed Transformers on many tasks, they lag behind Transformers on tasks which require strong copying or in-context learning abilities (e.g., 5-shot MMLU, Phonebook) or long-context reasoning. In contrast, we find that the 8B Mamba-2-Hybrid exceeds the 8B Transformer on all 12 standard tasks we evaluated (+2.65 points on average) and is predicted to be up to 8x faster when generating tokens at inference time. To validate long-context capabilities, we provide additional experiments evaluating variants of the Mamba-2-Hybrid and Transformer extended to support 16K, 32K, and 128K sequences. On an additional 23 long-context tasks, the hybrid model continues to closely match or exceed the Transformer on average. To enable further study, we release the checkpoints as well as the code used to train our models as part of NVIDIA's Megatron-LM project.
DiffiT: Diffusion Vision Transformers for Image Generation
Dec 04, 2023



Abstract:Diffusion models with their powerful expressivity and high sample quality have enabled many new applications and use-cases in various domains. For sample generation, these models rely on a denoising neural network that generates images by iterative denoising. Yet, the role of denoising network architecture is not well-studied with most efforts relying on convolutional residual U-Nets. In this paper, we study the effectiveness of vision transformers in diffusion-based generative learning. Specifically, we propose a new model, denoted as Diffusion Vision Transformers (DiffiT), which consists of a hybrid hierarchical architecture with a U-shaped encoder and decoder. We introduce a novel time-dependent self-attention module that allows attention layers to adapt their behavior at different stages of the denoising process in an efficient manner. We also introduce latent DiffiT which consists of transformer model with the proposed self-attention layers, for high-resolution image generation. Our results show that DiffiT is surprisingly effective in generating high-fidelity images, and it achieves state-of-the-art (SOTA) benchmarks on a variety of class-conditional and unconditional synthesis tasks. In the latent space, DiffiT achieves a new SOTA FID score of 1.73 on ImageNet-256 dataset. Repository: https://github.com/NVlabs/DiffiT
ViR: Vision Retention Networks
Oct 30, 2023Abstract:Vision Transformers (ViTs) have attracted a lot of popularity in recent years, due to their exceptional capabilities in modeling long-range spatial dependencies and scalability for large scale training. Although the training parallelism of self-attention mechanism plays an important role in retaining great performance, its quadratic complexity baffles the application of ViTs in many scenarios which demand fast inference. This effect is even more pronounced in applications in which autoregressive modeling of input features is required. In Natural Language Processing (NLP), a new stream of efforts have proposed parallelizable models with recurrent formulation that allows for efficient inference in generative applications. Inspired by this trend, we propose a new class of computer vision models, dubbed Vision Retention Networks (ViR), with dual parallel and recurrent formulations, which strike an optimal balance between fast inference and parallel training with competitive performance. In particular, ViR scales favorably for image throughput and memory consumption in tasks that require higher-resolution images due to its flexible formulation in processing large sequence lengths. The ViR is the first attempt to realize dual parallel and recurrent equivalency in a general vision backbone for recognition tasks. We have validated the effectiveness of ViR through extensive experiments with different dataset sizes and various image resolutions and achieved competitive performance. Our code and pretrained models will be made publicly available.
FasterViT: Fast Vision Transformers with Hierarchical Attention
Jun 09, 2023Abstract:We design a new family of hybrid CNN-ViT neural networks, named FasterViT, with a focus on high image throughput for computer vision (CV) applications. FasterViT combines the benefits of fast local representation learning in CNNs and global modeling properties in ViT. Our newly introduced Hierarchical Attention (HAT) approach decomposes global self-attention with quadratic complexity into a multi-level attention with reduced computational costs. We benefit from efficient window-based self-attention. Each window has access to dedicated carrier tokens that participate in local and global representation learning. At a high level, global self-attentions enable the efficient cross-window communication at lower costs. FasterViT achieves a SOTA Pareto-front in terms of accuracy \vs image throughput. We have extensively validated its effectiveness on various CV tasks including classification, object detection and segmentation. We also show that HAT can be used as a plug-and-play module for existing networks and enhance them. We further demonstrate significantly faster and more accurate performance than competitive counterparts for images with high resolution. Code is available at https://github.com/NVlabs/FasterViT.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Split-U-Net: Preventing Data Leakage in Split Learning for Collaborative Multi-Modal Brain Tumor Segmentation
Aug 22, 2022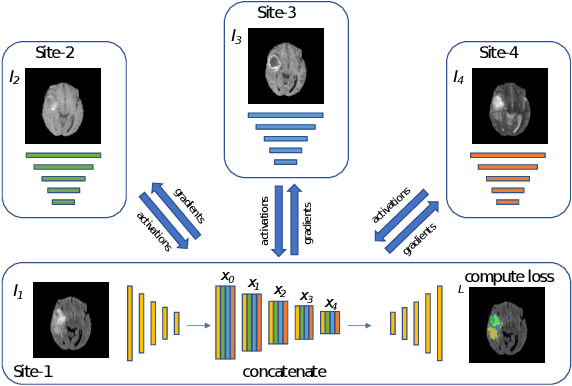
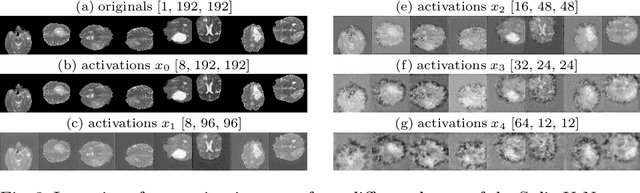
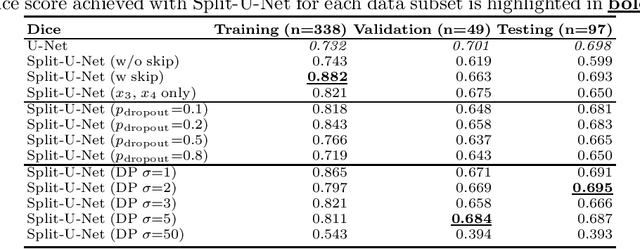
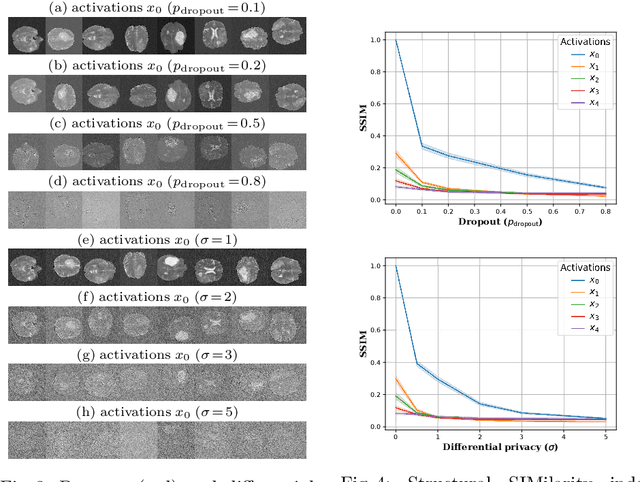
Abstract:Split learning (SL) has been proposed to train deep learning models in a decentralized manner. For decentralized healthcare applications with vertical data partitioning, SL can be beneficial as it allows institutes with complementary features or images for a shared set of patients to jointly develop more robust and generalizable models. In this work, we propose "Split-U-Net" and successfully apply SL for collaborative biomedical image segmentation. Nonetheless, SL requires the exchanging of intermediate activation maps and gradients to allow training models across different feature spaces, which might leak data and raise privacy concerns. Therefore, we also quantify the amount of data leakage in common SL scenarios for biomedical image segmentation and provide ways to counteract such leakage by applying appropriate defense strategies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge