Oliver Neumann
EAP4EMSIG -- Enhancing Event-Driven Microscopy for Microfluidic Single-Cell Analysis
Mar 30, 2025Abstract:Microfluidic Live-Cell Imaging yields data on microbial cell factories. However, continuous acquisition is challenging as high-throughput experiments often lack realtime insights, delaying responses to stochastic events. We introduce three components in the Experiment Automation Pipeline for Event-Driven Microscopy to Smart Microfluidic Single-Cell Analysis: a fast, accurate Deep Learning autofocusing method predicting the focus offset, an evaluation of real-time segmentation methods and a realtime data analysis dashboard. Our autofocusing achieves a Mean Absolute Error of 0.0226\textmu m with inference times below 50~ms. Among eleven Deep Learning segmentation methods, Cellpose~3 reached a Panoptic Quality of 93.58\%, while a distance-based method is fastest (121~ms, Panoptic Quality 93.02\%). All six Deep Learning Foundation Models were unsuitable for real-time segmentation.
Decision-Focused Fine-Tuning of Time Series Foundation Models for Dispatchable Feeder Optimization
Mar 03, 2025Abstract:Time series foundation models provide a universal solution for generating forecasts to support optimization problems in energy systems. Those foundation models are typically trained in a prediction-focused manner to maximize forecast quality. In contrast, decision-focused learning directly improves the resulting value of the forecast in downstream optimization rather than merely maximizing forecasting quality. The practical integration of forecast values into forecasting models is challenging, particularly when addressing complex applications with diverse instances, such as buildings. This becomes even more complicated when instances possess specific characteristics that require instance-specific, tailored predictions to increase the forecast value. To tackle this challenge, we use decision-focused fine-tuning within time series foundation models to offer a scalable and efficient solution for decision-focused learning applied to the dispatchable feeder optimization problem. To obtain more robust predictions for scarce building data, we use Moirai as a state-of-the-art foundation model, which offers robust and generalized results with few-shot parameter-efficient fine-tuning. Comparing the decision-focused fine-tuned Moirai with a state-of-the-art classical prediction-focused fine-tuning Morai, we observe an improvement of 9.45% in average total daily costs.
EAP4EMSIG -- Experiment Automation Pipeline for Event-Driven Microscopy to Smart Microfluidic Single-Cells Analysis
Nov 06, 2024Abstract:Microfluidic Live-Cell Imaging (MLCI) generates high-quality data that allows biotechnologists to study cellular growth dynamics in detail. However, obtaining these continuous data over extended periods is challenging, particularly in achieving accurate and consistent real-time event classification at the intersection of imaging and stochastic biology. To address this issue, we introduce the Experiment Automation Pipeline for Event-Driven Microscopy to Smart Microfluidic Single-Cells Analysis (EAP4EMSIG). In particular, we present initial zero-shot results from the real-time segmentation module of our approach. Our findings indicate that among four State-Of-The- Art (SOTA) segmentation methods evaluated, Omnipose delivers the highest Panoptic Quality (PQ) score of 0.9336, while Contour Proposal Network (CPN) achieves the fastest inference time of 185 ms with the second-highest PQ score of 0.8575. Furthermore, we observed that the vision foundation model Segment Anything is unsuitable for this particular use case.
AI-based automated active learning for discovery of hidden dynamic processes: A use case in light microscopy
Oct 05, 2023



Abstract:In the biomedical environment, experiments assessing dynamic processes are primarily performed by a human acquisition supervisor. Contemporary implementations of such experiments frequently aim to acquire a maximum number of relevant events from sometimes several hundred parallel, non-synchronous processes. Since in some high-throughput experiments, only one or a few instances of a given process can be observed simultaneously, a strategy for planning and executing an efficient acquisition paradigm is essential. To address this problem, we present two new methods in this paper. The first method, Encoded Dynamic Process (EDP), is Artificial Intelligence (AI)-based and represents dynamic processes so as to allow prediction of pseudo-time values from single still images. Second, with Experiment Automation Pipeline for Dynamic Processes (EAPDP), we present a Machine Learning Operations (MLOps)-based pipeline that uses the extracted knowledge from EDP to efficiently schedule acquisition in biomedical experiments for dynamic processes in practice. In a first experiment, we show that the pre-trained State-Of-The- Art (SOTA) object segmentation method Contour Proposal Networks (CPN) works reliably as a module of EAPDP to extract the relevant object for EDP from the acquired three-dimensional image stack.
MLOps for Scarce Image Data: A Use Case in Microscopic Image Analysis
Oct 04, 2023Abstract:Nowadays, Machine Learning (ML) is experiencing tremendous popularity that has never been seen before. The operationalization of ML models is governed by a set of concepts and methods referred to as Machine Learning Operations (MLOps). Nevertheless, researchers, as well as professionals, often focus more on the automation aspect and neglect the continuous deployment and monitoring aspects of MLOps. As a result, there is a lack of continuous learning through the flow of feedback from production to development, causing unexpected model deterioration over time due to concept drifts, particularly when dealing with scarce data. This work explores the complete application of MLOps in the context of scarce data analysis. The paper proposes a new holistic approach to enhance biomedical image analysis. Our method includes: a fingerprinting process that enables selecting the best models, datasets, and model development strategy relative to the image analysis task at hand; an automated model development stage; and a continuous deployment and monitoring process to ensure continuous learning. For preliminary results, we perform a proof of concept for fingerprinting in microscopic image datasets.
Transformer Training Strategies for Forecasting Multiple Load Time Series
Jun 19, 2023Abstract:Recent work uses Transformers for load forecasting, which are the state of the art for sequence modeling tasks in data-rich domains. In the smart grid of the future, accurate load forecasts must be provided on the level of individual clients of an energy supplier. While the total amount of electrical load data available to an energy supplier will increase with the ongoing smart meter rollout, the amount of data per client will always be limited. We test whether the Transformer benefits from a transfer learning strategy, where a global model is trained on the load time series data from multiple clients. We find that the global model is superior to two other training strategies commonly used in related work: multivariate models and local models. A comparison to linear models and multi-layer perceptrons shows that Transformers are effective for electrical load forecasting when they are trained with the right strategy.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
ProbPNN: Enhancing Deep Probabilistic Forecasting with Statistical Information
Feb 06, 2023



Abstract:Probabilistic forecasts are essential for various downstream applications such as business development, traffic planning, and electrical grid balancing. Many of these probabilistic forecasts are performed on time series data that contain calendar-driven periodicities. However, existing probabilistic forecasting methods do not explicitly take these periodicities into account. Therefore, in the present paper, we introduce a deep learning-based method that considers these calendar-driven periodicities explicitly. The present paper, thus, has a twofold contribution: First, we apply statistical methods that use calendar-driven prior knowledge to create rolling statistics and combine them with neural networks to provide better probabilistic forecasts. Second, we benchmark ProbPNN with state-of-the-art benchmarks by comparing the achieved normalised continuous ranked probability score (nCRPS) and normalised Pinball Loss (nPL) on two data sets containing in total more than 1000 time series. The results of the benchmarks show that using statistical forecasting components improves the probabilistic forecast performance and that ProbPNN outperforms other deep learning forecasting methods whilst requiring less computation costs.
EasyMLServe: Easy Deployment of REST Machine Learning Services
Nov 26, 2022



Abstract:Various research domains use machine learning approaches because they can solve complex tasks by learning from data. Deploying machine learning models, however, is not trivial and developers have to implement complete solutions which are often installed locally and include Graphical User Interfaces (GUIs). Distributing software to various users on-site has several problems. Therefore, we propose a concept to deploy software in the cloud. There are several frameworks available based on Representational State Transfer (REST) which can be used to implement cloud-based machine learning services. However, machine learning services for scientific users have special requirements that state-of-the-art REST frameworks do not cover completely. We contribute an EasyMLServe software framework to deploy machine learning services in the cloud using REST interfaces and generic local or web-based GUIs. Furthermore, we apply our framework on two real-world applications, \ie, energy time-series forecasting and cell instance segmentation. The EasyMLServe framework and the use cases are available on GitHub.
ciscNet -- A Single-Branch Cell Instance Segmentation and Classification Network
Feb 25, 2022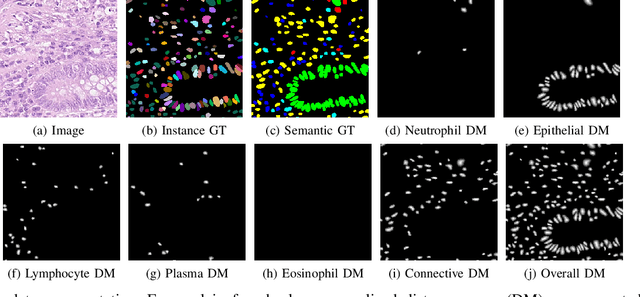
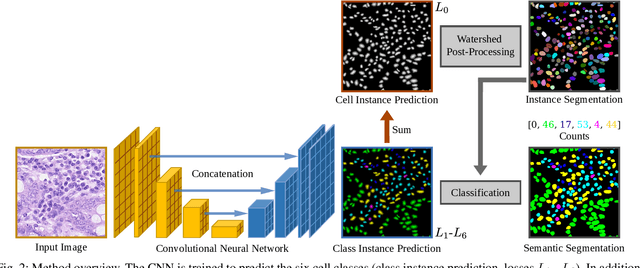
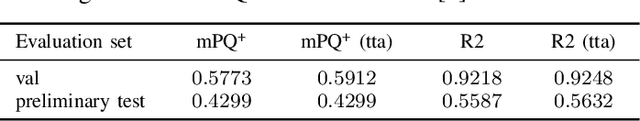
Abstract:Automated cell nucleus segmentation and classification are required to assist pathologists in their decision making. The Colon Nuclei Identification and Counting Challenge 2022 (CoNIC Challenge 2022) supports the development and comparability of segmentation and classification methods for histopathological images. In this contribution, we describe our CoNIC Challenge 2022 method ciscNet to segment, classify and count cell nuclei, and report preliminary evaluation results. Our code is available at https://git.scc.kit.edu/ciscnet/ciscnet-conic-2022.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge