Katharina Löffler
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
EmbedTrack -- Simultaneous Cell Segmentation and Tracking Through Learning Offsets and Clustering Bandwidths
Apr 25, 2022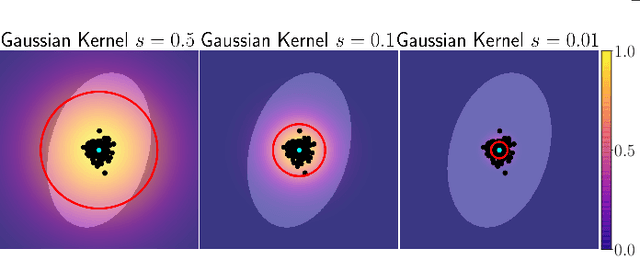

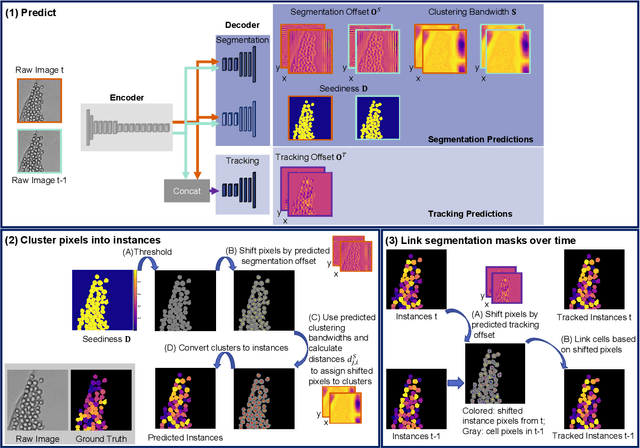
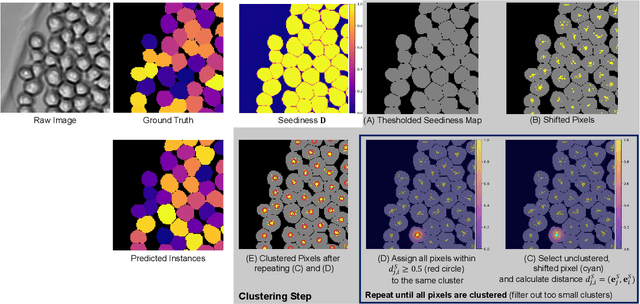
Abstract:A systematic analysis of the cell behavior requires automated approaches for cell segmentation and tracking. While deep learning has been successfully applied for the task of cell segmentation, there are few approaches for simultaneous cell segmentation and tracking using deep learning. Here, we present EmbedTrack, a single convolutional neural network for simultaneous cell segmentation and tracking which predicts easy to interpret embeddings. As embeddings, offsets of cell pixels to their cell center and bandwidths are learned. We benchmark our approach on nine 2D data sets from the Cell Tracking Challenge, where our approach performs on seven out of nine data sets within the top 3 contestants including three top 1 performances. The source code is publicly available at https://git.scc.kit.edu/kit-loe-ge/embedtrack.
ciscNet -- A Single-Branch Cell Instance Segmentation and Classification Network
Feb 25, 2022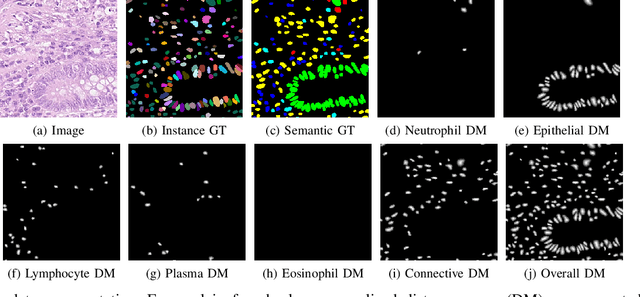
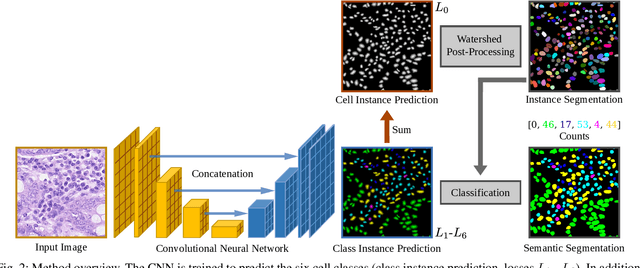
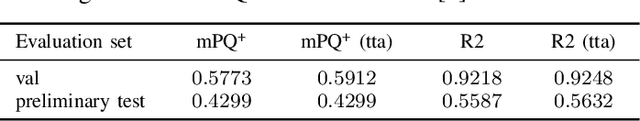
Abstract:Automated cell nucleus segmentation and classification are required to assist pathologists in their decision making. The Colon Nuclei Identification and Counting Challenge 2022 (CoNIC Challenge 2022) supports the development and comparability of segmentation and classification methods for histopathological images. In this contribution, we describe our CoNIC Challenge 2022 method ciscNet to segment, classify and count cell nuclei, and report preliminary evaluation results. Our code is available at https://git.scc.kit.edu/ciscnet/ciscnet-conic-2022.
Cell Segmentation and Tracking using Distance Transform Predictions and Movement Estimation with Graph-Based Matching
Apr 03, 2020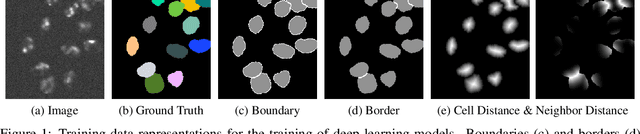

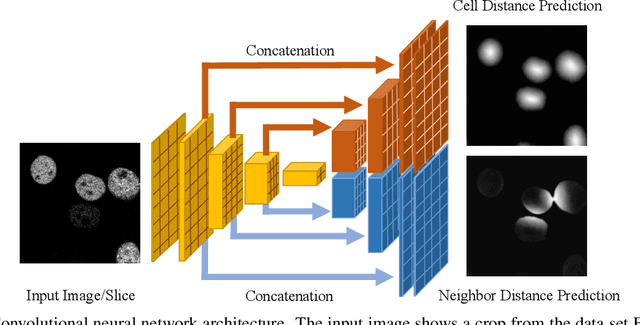
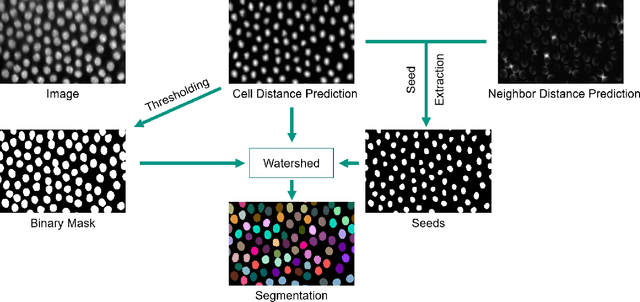
Abstract:In this paper, we present the approach used for our IEEE ISBI 2020 Cell Tracking Challenge contribution (team KIT-Sch-GE). Our method consists of a segmentation and a tracking step that includes the correction of segmentation errors (tracking by detection method). For the segmentation, deep learning-based predictions of cell distance maps and novel neighbor distance maps are used as input for a watershed post-processing. Since most of the provided Cell Tracking Challenge ground truth data are 2D, a 2D convolutional neural network is trained to predict the distance maps. The tracking is based on a movement estimation in combination with a matching formulated as a maximum flow minimum cost problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge