Marcel P. Schilling
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
ciscNet -- A Single-Branch Cell Instance Segmentation and Classification Network
Feb 25, 2022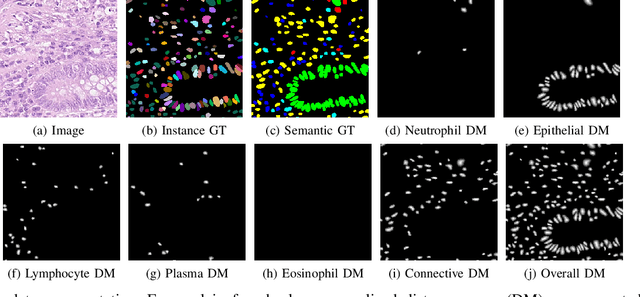
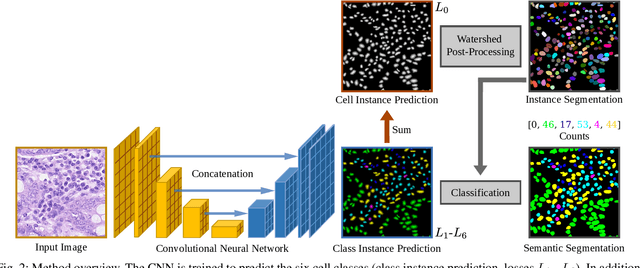
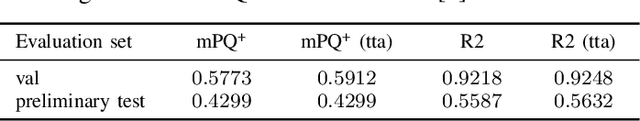
Abstract:Automated cell nucleus segmentation and classification are required to assist pathologists in their decision making. The Colon Nuclei Identification and Counting Challenge 2022 (CoNIC Challenge 2022) supports the development and comparability of segmentation and classification methods for histopathological images. In this contribution, we describe our CoNIC Challenge 2022 method ciscNet to segment, classify and count cell nuclei, and report preliminary evaluation results. Our code is available at https://git.scc.kit.edu/ciscnet/ciscnet-conic-2022.
Label Assistant: A Workflow for Assisted Data Annotation in Image Segmentation Tasks
Nov 27, 2021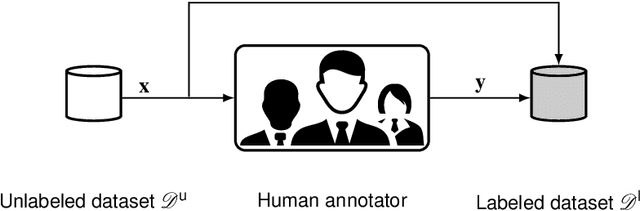
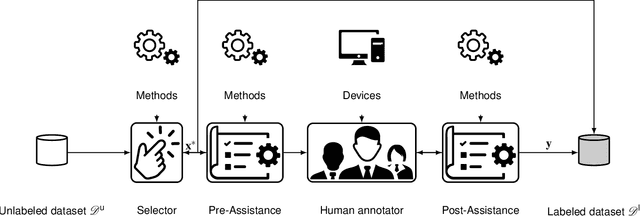
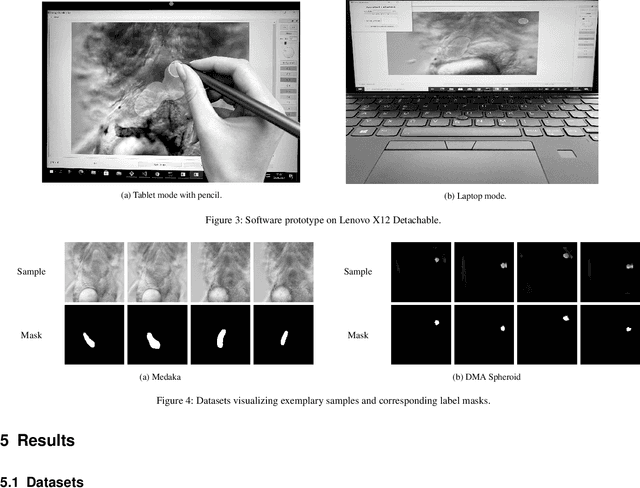
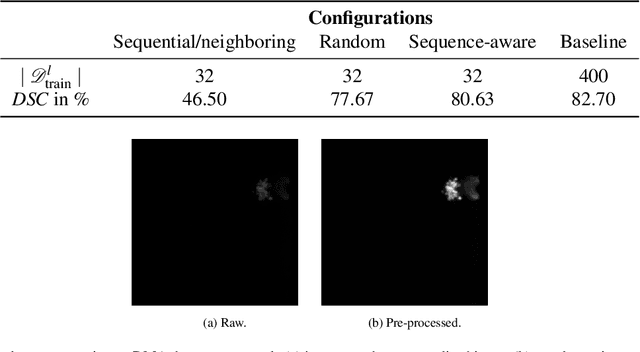
Abstract:Recent research in the field of computer vision strongly focuses on deep learning architectures to tackle image processing problems. Deep neural networks are often considered in complex image processing scenarios since traditional computer vision approaches are expensive to develop or reach their limits due to complex relations. However, a common criticism is the need for large annotated datasets to determine robust parameters. Annotating images by human experts is time-consuming, burdensome, and expensive. Thus, support is needed to simplify annotation, increase user efficiency, and annotation quality. In this paper, we propose a generic workflow to assist the annotation process and discuss methods on an abstract level. Thereby, we review the possibilities of focusing on promising samples, image pre-processing, pre-labeling, label inspection, or post-processing of annotations. In addition, we present an implementation of the proposal by means of a developed flexible and extendable software prototype nested in hybrid touchscreen/laptop device.
Radar Artifact Labeling Framework (RALF): Method for Plausible Radar Detections in Datasets
Dec 03, 2020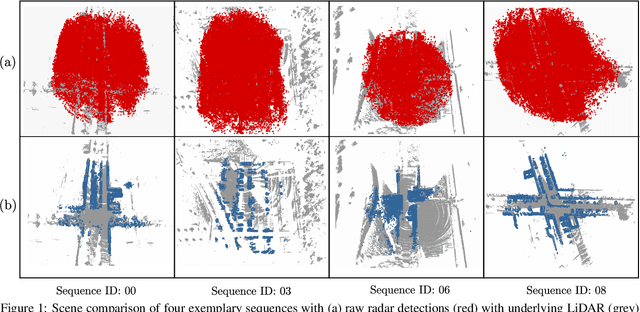
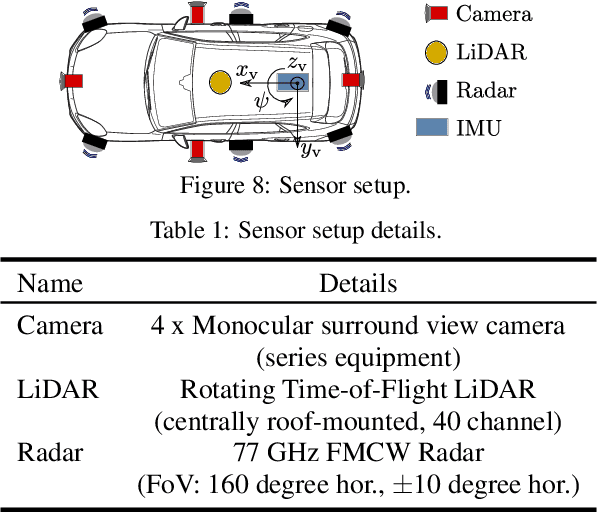
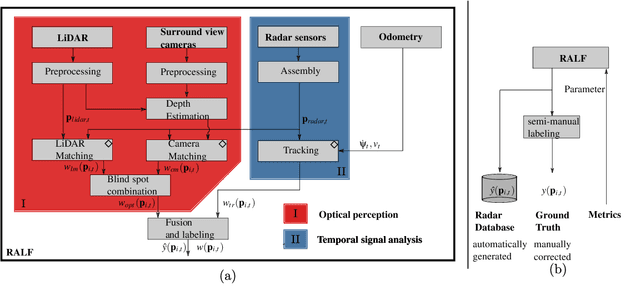
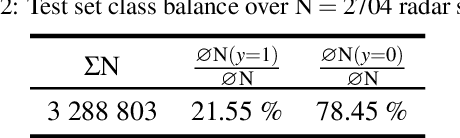
Abstract:Research on localization and perception for Autonomous Driving is mainly focused on camera and LiDAR datasets, rarely on radar data. Manually labeling sparse radar point clouds is challenging. For a dataset generation, we propose the cross sensor Radar Artifact Labeling Framework (RALF). Automatically generated labels for automotive radar data help to cure radar shortcomings like artifacts for the application of artificial intelligence. RALF provides plausibility labels for radar raw detections, distinguishing between artifacts and targets. The optical evaluation backbone consists of a generalized monocular depth image estimation of surround view cameras plus LiDAR scans. Modern car sensor sets of cameras and LiDAR allow to calibrate image-based relative depth information in overlapping sensing areas. K-Nearest Neighbors matching relates the optical perception point cloud with raw radar detections. In parallel, a temporal tracking evaluation part considers the radar detections' transient behavior. Based on the distance between matches, respecting both sensor and model uncertainties, we propose a plausibility rating of every radar detection. We validate the results by evaluating error metrics on semi-manually labeled ground truth dataset of $3.28\cdot10^6$ points. Besides generating plausible radar detections, the framework enables further labeled low-level radar signal datasets for applications of perception and Autonomous Driving learning tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge