Mohammad Yaqub
FUGC: Benchmarking Semi-Supervised Learning Methods for Cervical Segmentation
Jan 22, 2026Abstract:Accurate segmentation of cervical structures in transvaginal ultrasound (TVS) is critical for assessing the risk of spontaneous preterm birth (PTB), yet the scarcity of labeled data limits the performance of supervised learning approaches. This paper introduces the Fetal Ultrasound Grand Challenge (FUGC), the first benchmark for semi-supervised learning in cervical segmentation, hosted at ISBI 2025. FUGC provides a dataset of 890 TVS images, including 500 training images, 90 validation images, and 300 test images. Methods were evaluated using the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), and runtime (RT), with a weighted combination of 0.4/0.4/0.2. The challenge attracted 10 teams with 82 participants submitting innovative solutions. The best-performing methods for each individual metric achieved 90.26\% mDSC, 38.88 mHD, and 32.85 ms RT, respectively. FUGC establishes a standardized benchmark for cervical segmentation, demonstrates the efficacy of semi-supervised methods with limited labeled data, and provides a foundation for AI-assisted clinical PTB risk assessment.
FETAL-GAUGE: A Benchmark for Assessing Vision-Language Models in Fetal Ultrasound
Dec 25, 2025Abstract:The growing demand for prenatal ultrasound imaging has intensified a global shortage of trained sonographers, creating barriers to essential fetal health monitoring. Deep learning has the potential to enhance sonographers' efficiency and support the training of new practitioners. Vision-Language Models (VLMs) are particularly promising for ultrasound interpretation, as they can jointly process images and text to perform multiple clinical tasks within a single framework. However, despite the expansion of VLMs, no standardized benchmark exists to evaluate their performance in fetal ultrasound imaging. This gap is primarily due to the modality's challenging nature, operator dependency, and the limited public availability of datasets. To address this gap, we present Fetal-Gauge, the first and largest visual question answering benchmark specifically designed to evaluate VLMs across various fetal ultrasound tasks. Our benchmark comprises over 42,000 images and 93,000 question-answer pairs, spanning anatomical plane identification, visual grounding of anatomical structures, fetal orientation assessment, clinical view conformity, and clinical diagnosis. We systematically evaluate several state-of-the-art VLMs, including general-purpose and medical-specific models, and reveal a substantial performance gap: the best-performing model achieves only 55\% accuracy, far below clinical requirements. Our analysis identifies critical limitations of current VLMs in fetal ultrasound interpretation, highlighting the urgent need for domain-adapted architectures and specialized training approaches. Fetal-Gauge establishes a rigorous foundation for advancing multimodal deep learning in prenatal care and provides a pathway toward addressing global healthcare accessibility challenges. Our benchmark will be publicly available once the paper gets accepted.
MMRINet: Efficient Mamba-Based Segmentation with Dual-Path Refinement for Low-Resource MRI Analysis
Nov 15, 2025Abstract:Automated brain tumor segmentation in multi-parametric MRI remains challenging in resource-constrained settings where deep 3D networks are computationally prohibitive. We propose MMRINet, a lightweight architecture that replaces quadratic-complexity attention with linear-complexity Mamba state-space models for efficient volumetric context modeling. Novel Dual-Path Feature Refinement (DPFR) modules maximize feature diversity without additional data requirements, while Progressive Feature Aggregation (PFA) enables effective multi-scale fusion. In the BraTS-Lighthouse SSA 2025, our model achieves strong performance with an average Dice score of (0.752) and an average HD95 of (12.23) with only ~2.5M parameters, demonstrating efficient and accurate segmentation suitable for low-resource clinical environments. Our GitHub repository can be accessed here: github.com/BioMedIA-MBZUAI/MMRINet.
MAFM^3: Modular Adaptation of Foundation Models for Multi-Modal Medical AI
Nov 14, 2025Abstract:Foundational models are trained on extensive datasets to capture the general trends of a domain. However, in medical imaging, the scarcity of data makes pre-training for every domain, modality, or task challenging. Instead of building separate models, we propose MAFM^3 (Modular Adaptation of Foundation Models for Multi-Modal Medical AI), a framework that enables a single foundation model to expand into diverse domains, tasks, and modalities through lightweight modular components. These components serve as specialized skill sets that allow the system to flexibly activate the appropriate capability at the inference time, depending on the input type or clinical objective. Unlike conventional adaptation methods that treat each new task or modality in isolation, MAFM^3 provides a unified and expandable framework for efficient multitask and multimodality adaptation. Empirically, we validate our approach by adapting a chest CT foundation model initially trained for classification into prognosis and segmentation modules. Our results show improved performance on both tasks. Furthermore, by incorporating PET scans, MAFM^3 achieved an improvement in the Dice score 5% compared to the respective baselines. These findings establish that foundation models, when equipped with modular components, are not inherently constrained to their initial training scope but can evolve into multitask, multimodality systems for medical imaging. The code implementation of this work can be found at https://github.com/Areeb2735/CTscan_prognosis_VLM
T3: Test-Time Model Merging in VLMs for Zero-Shot Medical Imaging Analysis
Oct 31, 2025



Abstract:In medical imaging, vision-language models face a critical duality: pretrained networks offer broad robustness but lack subtle, modality-specific characteristics, while fine-tuned expert models achieve high in-distribution accuracy yet falter under modality shift. Existing model-merging techniques, designed for natural-image benchmarks, are simple and efficient but fail to deliver consistent gains across diverse medical modalities; their static interpolation limits reliability in varied clinical tasks. To address this, we introduce Test-Time Task adaptive merging (T^3), a backpropagation-free framework that computes per-sample interpolation coefficients via the Jensen-Shannon divergence between the two models' output distributions. T^3 dynamically preserves local precision when models agree and defers to generalist robustness under drift. To overcome the inference costs of sample-wise merging, we further propose a batch-wise extension, T^3_B, that computes a merging coefficient across a batch of samples, dramatically reducing computational bottleneck. Recognizing the lack of a standardized medical-merging benchmark, we present a rigorous cross-evaluation protocol spanning in-domain, base-to-novel, and corruptions across four modalities. Empirically, T^3 sets new state-of-the-art in Top-1 accuracy and error reduction, outperforming strong baselines while maintaining efficiency, paving the way for adaptive MVLM deployment in clinical settings. Our code is available at https://github.com/Razaimam45/TCube.
BRIQA: Balanced Reweighting in Image Quality Assessment of Pediatric Brain MRI
Oct 30, 2025Abstract:Assessing the severity of artifacts in pediatric brain Magnetic Resonance Imaging (MRI) is critical for diagnostic accuracy, especially in low-field systems where the signal-to-noise ratio is reduced. Manual quality assessment is time-consuming and subjective, motivating the need for robust automated solutions. In this work, we propose BRIQA (Balanced Reweighting in Image Quality Assessment), which addresses class imbalance in artifact severity levels. BRIQA uses gradient-based loss reweighting to dynamically adjust per-class contributions and employs a rotating batching scheme to ensure consistent exposure to underrepresented classes. Through experiments, no single architecture performs best across all artifact types, emphasizing the importance of architectural diversity. The rotating batching configuration improves performance across metrics by promoting balanced learning when combined with cross-entropy loss. BRIQA improves average macro F1 score from 0.659 to 0.706, with notable gains in Noise (0.430), Zipper (0.098), Positioning (0.097), Contrast (0.217), Motion (0.022), and Banding (0.012) artifact severity classification. The code is available at https://github.com/BioMedIA-MBZUAI/BRIQA.
CardioBench: Do Echocardiography Foundation Models Generalize Beyond the Lab?
Oct 01, 2025Abstract:Foundation models (FMs) are reshaping medical imaging, yet their application in echocardiography remains limited. While several echocardiography-specific FMs have recently been introduced, no standardized benchmark exists to evaluate them. Echocardiography poses unique challenges, including noisy acquisitions, high frame redundancy, and limited public datasets. Most existing solutions evaluate on private data, restricting comparability. To address this, we introduce CardioBench, a comprehensive benchmark for echocardiography FMs. CardioBench unifies eight publicly available datasets into a standardized suite spanning four regression and five classification tasks, covering functional, structural, diagnostic, and view recognition endpoints. We evaluate several leading FM, including cardiac-specific, biomedical, and general-purpose encoders, under consistent zero-shot, probing, and alignment protocols. Our results highlight complementary strengths across model families: temporal modeling is critical for functional regression, retrieval provides robustness under distribution shift, and domain-specific text encoders capture physiologically meaningful axes. General-purpose encoders transfer strongly and often close the gap with probing, but struggle with fine-grained distinctions like view classification and subtle pathology recognition. By releasing preprocessing, splits, and public evaluation pipelines, CardioBench establishes a reproducible reference point and offers actionable insights to guide the design of future echocardiography foundation models.
Decoupling Clinical and Class-Agnostic Features for Reliable Few-Shot Adaptation under Shift
Sep 11, 2025Abstract:Medical vision-language models (VLMs) offer promise for clinical decision support, yet their reliability under distribution shifts remains a major concern for safe deployment. These models often learn task-agnostic correlations due to variability in imaging protocols and free-text reports, limiting their generalizability and increasing the risk of failure in real-world settings. We propose DRiFt, a structured feature decoupling framework that explicitly separates clinically relevant signals from task-agnostic noise using parameter-efficient tuning (LoRA) and learnable prompt tokens. To enhance cross-modal alignment and reduce uncertainty, we curate high-quality, clinically grounded image-text pairs by generating captions for a diverse medical dataset. Our approach improves in-distribution performance by +11.4% Top-1 accuracy and +3.3% Macro-F1 over prior prompt-based methods, while maintaining strong robustness across unseen datasets. Ablation studies reveal that disentangling task-relevant features and careful alignment significantly enhance model generalization and reduce unpredictable behavior under domain shift. These insights contribute toward building safer, more trustworthy VLMs for clinical use. The code is available at https://github.com/rumaima/DRiFt.
Advancing Fetal Ultrasound Image Quality Assessment in Low-Resource Settings
Jul 30, 2025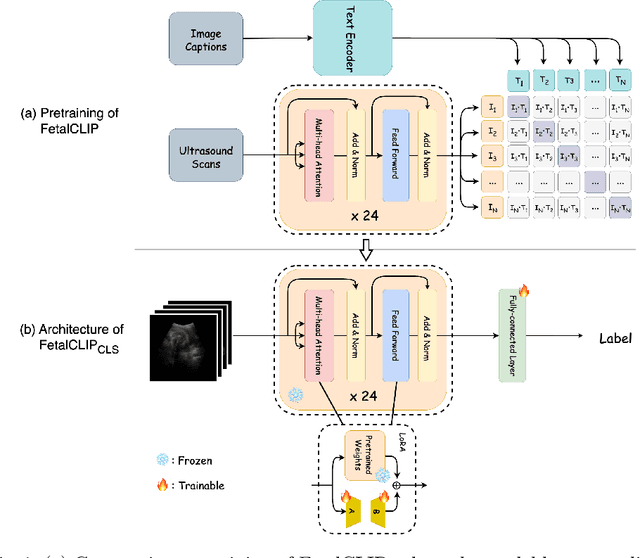
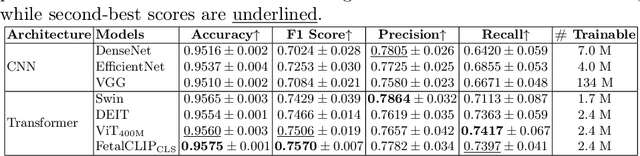
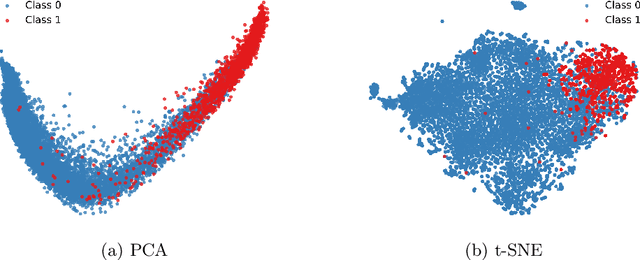

Abstract:Accurate fetal biometric measurements, such as abdominal circumference, play a vital role in prenatal care. However, obtaining high-quality ultrasound images for these measurements heavily depends on the expertise of sonographers, posing a significant challenge in low-income countries due to the scarcity of trained personnel. To address this issue, we leverage FetalCLIP, a vision-language model pretrained on a curated dataset of over 210,000 fetal ultrasound image-caption pairs, to perform automated fetal ultrasound image quality assessment (IQA) on blind-sweep ultrasound data. We introduce FetalCLIP$_{CLS}$, an IQA model adapted from FetalCLIP using Low-Rank Adaptation (LoRA), and evaluate it on the ACOUSLIC-AI dataset against six CNN and Transformer baselines. FetalCLIP$_{CLS}$ achieves the highest F1 score of 0.757. Moreover, we show that an adapted segmentation model, when repurposed for classification, further improves performance, achieving an F1 score of 0.771. Our work demonstrates how parameter-efficient fine-tuning of fetal ultrasound foundation models can enable task-specific adaptations, advancing prenatal care in resource-limited settings. The experimental code is available at: https://github.com/donglihe-hub/FetalCLIP-IQA.
Not Only Grey Matter: OmniBrain for Robust Multimodal Classification of Alzheimer's Disease
Jul 28, 2025



Abstract:Alzheimer's disease affects over 55 million people worldwide and is projected to more than double by 2050, necessitating rapid, accurate, and scalable diagnostics. However, existing approaches are limited because they cannot achieve clinically acceptable accuracy, generalization across datasets, robustness to missing modalities, and explainability all at the same time. This inability to satisfy all these requirements simultaneously undermines their reliability in clinical settings. We propose OmniBrain, a multimodal framework that integrates brain MRI, radiomics, gene expression, and clinical data using a unified model with cross-attention and modality dropout. OmniBrain achieves $92.2 \pm 2.4\%$accuracy on the ANMerge dataset and generalizes to the MRI-only ADNI dataset with $70.4 \pm 2.7\%$ accuracy, outperforming unimodal and prior multimodal approaches. Explainability analyses highlight neuropathologically relevant brain regions and genes, enhancing clinical trust. OmniBrain offers a robust, interpretable, and practical solution for real-world Alzheimer's diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge