Christian F. Baumgartner
A Critical Perspective on Finite Sample Conformal Prediction Theory in Medical Applications
Dec 09, 2025Abstract:Machine learning (ML) is transforming healthcare, but safe clinical decisions demand reliable uncertainty estimates that standard ML models fail to provide. Conformal prediction (CP) is a popular tool that allows users to turn heuristic uncertainty estimates into uncertainty estimates with statistical guarantees. CP works by converting predictions of a ML model, together with a calibration sample, into prediction sets that are guaranteed to contain the true label with any desired probability. An often cited advantage is that CP theory holds for calibration samples of arbitrary size, suggesting that uncertainty estimates with practically meaningful statistical guarantees can be achieved even if only small calibration sets are available. We question this promise by showing that, although the statistical guarantees hold for calibration sets of arbitrary size, the practical utility of these guarantees does highly depend on the size of the calibration set. This observation is relevant in medical domains because data is often scarce and obtaining large calibration sets is therefore infeasible. We corroborate our critique in an empirical demonstration on a medical image classification task.
Understanding Benefits and Pitfalls of Current Methods for the Segmentation of Undersampled MRI Data
Aug 26, 2025Abstract:MR imaging is a valuable diagnostic tool allowing to non-invasively visualize patient anatomy and pathology with high soft-tissue contrast. However, MRI acquisition is typically time-consuming, leading to patient discomfort and increased costs to the healthcare system. Recent years have seen substantial research effort into the development of methods that allow for accelerated MRI acquisition while still obtaining a reconstruction that appears similar to the fully-sampled MR image. However, for many applications a perfectly reconstructed MR image may not be necessary, particularly, when the primary goal is a downstream task such as segmentation. This has led to growing interest in methods that aim to perform segmentation directly on accelerated MRI data. Despite recent advances, existing methods have largely been developed in isolation, without direct comparison to one another, often using separate or private datasets, and lacking unified evaluation standards. To date, no high-quality, comprehensive comparison of these methods exists, and the optimal strategy for segmenting accelerated MR data remains unknown. This paper provides the first unified benchmark for the segmentation of undersampled MRI data comparing 7 approaches. A particular focus is placed on comparing \textit{one-stage approaches}, that combine reconstruction and segmentation into a unified model, with \textit{two-stage approaches}, that utilize established MRI reconstruction methods followed by a segmentation network. We test these methods on two MRI datasets that include multi-coil k-space data as well as a human-annotated segmentation ground-truth. We find that simple two-stage methods that consider data-consistency lead to the best segmentation scores, surpassing complex specialized methods that are developed specifically for this task.
MRExtrap: Longitudinal Aging of Brain MRIs using Linear Modeling in Latent Space
Aug 26, 2025Abstract:Simulating aging in 3D brain MRI scans can reveal disease progression patterns in neurological disorders such as Alzheimer's disease. Current deep learning-based generative models typically approach this problem by predicting future scans from a single observed scan. We investigate modeling brain aging via linear models in the latent space of convolutional autoencoders (MRExtrap). Our approach, MRExtrap, is based on our observation that autoencoders trained on brain MRIs create latent spaces where aging trajectories appear approximately linear. We train autoencoders on brain MRIs to create latent spaces, and investigate how these latent spaces allow predicting future MRIs through linear extrapolation based on age, using an estimated latent progression rate $\boldsymbol{\beta}$. For single-scan prediction, we propose using population-averaged and subject-specific priors on linear progression rates. We also demonstrate that predictions in the presence of additional scans can be flexibly updated using Bayesian posterior sampling, providing a mechanism for subject-specific refinement. On the ADNI dataset, MRExtrap predicts aging patterns accurately and beats a GAN-based baseline for single-volume prediction of brain aging. We also demonstrate and analyze multi-scan conditioning to incorporate subject-specific progression rates. Finally, we show that the latent progression rates in MRExtrap's linear framework correlate with disease and age-based aging patterns from previously studied structural atrophy rates. MRExtrap offers a simple and robust method for the age-based generation of 3D brain MRIs, particularly valuable in scenarios with multiple longitudinal observations.
Subgroup Performance Analysis in Hidden Stratifications
Mar 13, 2025Abstract:Machine learning (ML) models may suffer from significant performance disparities between patient groups. Identifying such disparities by monitoring performance at a granular level is crucial for safely deploying ML to each patient. Traditional subgroup analysis based on metadata can expose performance disparities only if the available metadata (e.g., patient sex) sufficiently reflects the main reasons for performance variability, which is not common. Subgroup discovery techniques that identify cohesive subgroups based on learned feature representations appear as a potential solution: They could expose hidden stratifications and provide more granular subgroup performance reports. However, subgroup discovery is challenging to evaluate even as a standalone task, as ground truth stratification labels do not exist in real data. Subgroup discovery has thus neither been applied nor evaluated for the application of subgroup performance monitoring. Here, we apply subgroup discovery for performance monitoring in chest x-ray and skin lesion classification. We propose novel evaluation strategies and show that a simplified subgroup discovery method without access to classification labels or metadata can expose larger performance disparities than traditional metadata-based subgroup analysis. We provide the first compelling evidence that subgroup discovery can serve as an important tool for comprehensive performance validation and monitoring of trustworthy AI in medicine.
Prototype-Based Multiple Instance Learning for Gigapixel Whole Slide Image Classification
Mar 11, 2025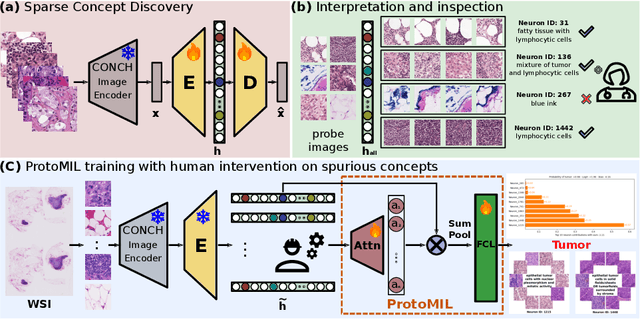
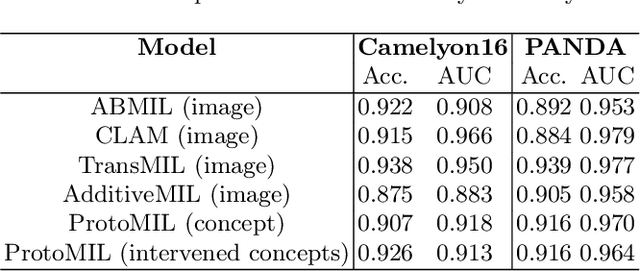
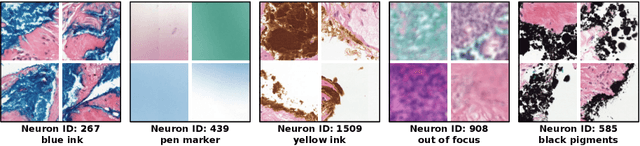
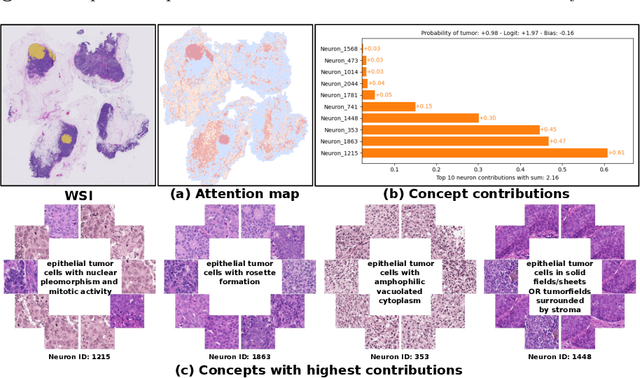
Abstract:Multiple Instance Learning (MIL) methods have succeeded remarkably in histopathology whole slide image (WSI) analysis. However, most MIL models only offer attention-based explanations that do not faithfully capture the model's decision mechanism and do not allow human-model interaction. To address these limitations, we introduce ProtoMIL, an inherently interpretable MIL model for WSI analysis that offers user-friendly explanations and supports human intervention. Our approach employs a sparse autoencoder to discover human-interpretable concepts from the image feature space, which are then used to train ProtoMIL. The model represents predictions as linear combinations of concepts, making the decision process transparent. Furthermore, ProtoMIL allows users to perform model interventions by altering the input concepts. Experiments on two widely used pathology datasets demonstrate that ProtoMIL achieves a classification performance comparable to state-of-the-art MIL models while offering intuitively understandable explanations. Moreover, we demonstrate that our method can eliminate reliance on diagnostically irrelevant information via human intervention, guiding the model toward being right for the right reason. Code will be publicly available at https://github.com/ss-sun/ProtoMIL.
Label-free Concept Based Multiple Instance Learning for Gigapixel Histopathology
Jan 06, 2025



Abstract:Multiple Instance Learning (MIL) methods allow for gigapixel Whole-Slide Image (WSI) analysis with only slide-level annotations. Interpretability is crucial for safely deploying such algorithms in high-stakes medical domains. Traditional MIL methods offer explanations by highlighting salient regions. However, such spatial heatmaps provide limited insights for end users. To address this, we propose a novel inherently interpretable WSI-classification approach that uses human-understandable pathology concepts to generate explanations. Our proposed Concept MIL model leverages recent advances in vision-language models to directly predict pathology concepts based on image features. The model's predictions are obtained through a linear combination of the concepts identified on the top-K patches of a WSI, enabling inherent explanations by tracing each concept's influence on the prediction. In contrast to traditional concept-based interpretable models, our approach eliminates the need for costly human annotations by leveraging the vision-language model. We validate our method on two widely used pathology datasets: Camelyon16 and PANDA. On both datasets, Concept MIL achieves AUC and accuracy scores over 0.9, putting it on par with state-of-the-art models. We further find that 87.1\% (Camelyon16) and 85.3\% (PANDA) of the top 20 patches fall within the tumor region. A user study shows that the concepts identified by our model align with the concepts used by pathologists, making it a promising strategy for human-interpretable WSI classification.
Navigating Data Scarcity using Foundation Models: A Benchmark of Few-Shot and Zero-Shot Learning Approaches in Medical Imaging
Aug 15, 2024Abstract:Data scarcity is a major limiting factor for applying modern machine learning techniques to clinical tasks. Although sufficient data exists for some well-studied medical tasks, there remains a long tail of clinically relevant tasks with poor data availability. Recently, numerous foundation models have demonstrated high suitability for few-shot learning (FSL) and zero-shot learning (ZSL), potentially making them more accessible to practitioners. However, it remains unclear which foundation model performs best on FSL medical image analysis tasks and what the optimal methods are for learning from limited data. We conducted a comprehensive benchmark study of ZSL and FSL using 16 pretrained foundation models on 19 diverse medical imaging datasets. Our results indicate that BiomedCLIP, a model pretrained exclusively on medical data, performs best on average for very small training set sizes, while very large CLIP models pretrained on LAION-2B perform best with slightly more training samples. However, simply fine-tuning a ResNet-18 pretrained on ImageNet performs similarly with more than five training examples per class. Our findings also highlight the need for further research on foundation models specifically tailored for medical applications and the collection of more datasets to train these models.
Segmentation-guided MRI reconstruction for meaningfully diverse reconstructions
Jul 25, 2024



Abstract:Inverse problems, such as accelerated MRI reconstruction, are ill-posed and an infinite amount of possible and plausible solutions exist. This may not only lead to uncertainty in the reconstructed image but also in downstream tasks such as semantic segmentation. This uncertainty, however, is mostly not analyzed in the literature, even though probabilistic reconstruction models are commonly used. These models can be prone to ignore plausible but unlikely solutions like rare pathologies. Building on MRI reconstruction approaches based on diffusion models, we add guidance to the diffusion process during inference, generating two meaningfully diverse reconstructions corresponding to an upper and lower bound segmentation. The reconstruction uncertainty can then be quantified by the difference between these bounds, which we coin the 'uncertainty boundary'. We analyzed the behavior of the upper and lower bound segmentations for a wide range of acceleration factors and found the uncertainty boundary to be both more reliable and more accurate compared to repeated sampling. Code is available at https://github.com/NikolasMorshuis/SGR
Conformal Performance Range Prediction for Segmentation Output Quality Control
Jul 18, 2024



Abstract:Recent works have introduced methods to estimate segmentation performance without ground truth, relying solely on neural network softmax outputs. These techniques hold potential for intuitive output quality control. However, such performance estimates rely on calibrated softmax outputs, which is often not the case in modern neural networks. Moreover, the estimates do not take into account inherent uncertainty in segmentation tasks. These limitations may render precise performance predictions unattainable, restricting the practical applicability of performance estimation methods. To address these challenges, we develop a novel approach for predicting performance ranges with statistical guarantees of containing the ground truth with a user specified probability. Our method leverages sampling-based segmentation uncertainty estimation to derive heuristic performance ranges, and applies split conformal prediction to transform these estimates into rigorous prediction ranges that meet the desired guarantees. We demonstrate our approach on the FIVES retinal vessel segmentation dataset and compare five commonly used sampling-based uncertainty estimation techniques. Our results show that it is possible to achieve the desired coverage with small prediction ranges, highlighting the potential of performance range prediction as a valuable tool for output quality control.
PULPo: Probabilistic Unsupervised Laplacian Pyramid Registration
Jul 15, 2024



Abstract:Deformable image registration is fundamental to many medical imaging applications. Registration is an inherently ambiguous task often admitting many viable solutions. While neural network-based registration techniques enable fast and accurate registration, the majority of existing approaches are not able to estimate uncertainty. Here, we present PULPo, a method for probabilistic deformable registration capable of uncertainty quantification. PULPo probabilistically models the distribution of deformation fields on different hierarchical levels combining them using Laplacian pyramids. This allows our method to model global as well as local aspects of the deformation field. We evaluate our method on two widely used neuroimaging datasets and find that it achieves high registration performance as well as substantially better calibrated uncertainty quantification compared to the current state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge