Michiel Schaap
Vector Representations of Vessel Trees
Jun 11, 2025Abstract:We introduce a novel framework for learning vector representations of tree-structured geometric data focusing on 3D vascular networks. Our approach employs two sequentially trained Transformer-based autoencoders. In the first stage, the Vessel Autoencoder captures continuous geometric details of individual vessel segments by learning embeddings from sampled points along each curve. In the second stage, the Vessel Tree Autoencoder encodes the topology of the vascular network as a single vector representation, leveraging the segment-level embeddings from the first model. A recursive decoding process ensures that the reconstructed topology is a valid tree structure. Compared to 3D convolutional models, this proposed approach substantially lowers GPU memory requirements, facilitating large-scale training. Experimental results on a 2D synthetic tree dataset and a 3D coronary artery dataset demonstrate superior reconstruction fidelity, accurate topology preservation, and realistic interpolations in latent space. Our scalable framework, named VeTTA, offers precise, flexible, and topologically consistent modeling of anatomical tree structures in medical imaging.
A Diffusion Model for Simulation Ready Coronary Anatomy with Morpho-skeletal Control
Jul 23, 2024



Abstract:Virtual interventions enable the physics-based simulation of device deployment within coronary arteries. This framework allows for counterfactual reasoning by deploying the same device in different arterial anatomies. However, current methods to create such counterfactual arteries face a trade-off between controllability and realism. In this study, we investigate how Latent Diffusion Models (LDMs) can custom synthesize coronary anatomy for virtual intervention studies based on mid-level anatomic constraints such as topological validity, local morphological shape, and global skeletal structure. We also extend diffusion model guidance strategies to the context of morpho-skeletal conditioning and propose a novel guidance method for continuous attributes that adaptively updates the negative guiding condition throughout sampling. Our framework enables the generation and editing of coronary anatomy in a controllable manner, allowing device designers to derive mechanistic insights regarding anatomic variation and simulated device deployment.
Image To Tree with Recursive Prompting
Jan 01, 2023



Abstract:Extracting complex structures from grid-based data is a common key step in automated medical image analysis. The conventional solution to recovering tree-structured geometries typically involves computing the minimal cost path through intermediate representations derived from segmentation masks. However, this methodology has significant limitations in the context of projective imaging of tree-structured 3D anatomical data such as coronary arteries, since there are often overlapping branches in the 2D projection. In this work, we propose a novel approach to predicting tree connectivity structure which reformulates the task as an optimization problem over individual steps of a recursive process. We design and train a two-stage model which leverages the UNet and Transformer architectures and introduces an image-based prompting technique. Our proposed method achieves compelling results on a pair of synthetic datasets, and outperforms a shortest-path baseline.
Morphology-based non-rigid registration of coronary computed tomography and intravascular images through virtual catheter path optimization
Dec 30, 2022



Abstract:Coronary Computed Tomography Angiography (CCTA) provides information on the presence, extent, and severity of obstructive coronary artery disease. Large-scale clinical studies analyzing CCTA-derived metrics typically require ground-truth validation in the form of high-fidelity 3D intravascular imaging. However, manual rigid alignment of intravascular images to corresponding CCTA images is both time consuming and user-dependent. Moreover, intravascular modalities suffer from several non-rigid motion-induced distortions arising from distortions in the imaging catheter path. To address these issues, we here present a semi-automatic segmentation-based framework for both rigid and non-rigid matching of intravascular images to CCTA images. We formulate the problem in terms of finding the optimal \emph{virtual catheter path} that samples the CCTA data to recapitulate the coronary artery morphology found in the intravascular image. We validate our co-registration framework on a cohort of $n=40$ patients using bifurcation landmarks as ground truth for longitudinal and rotational registration. Our results indicate that our non-rigid registration significantly outperforms other co-registration approaches for luminal bifurcation alignment in both longitudinal (mean mismatch: 3.3 frames) and rotational directions (mean mismatch: 28.6 degrees). By providing a differentiable framework for automatic multi-modal intravascular data fusion, our developed co-registration modules significantly reduces the manual effort required to conduct large-scale multi-modal clinical studies while also providing a solid foundation for the development of machine learning-based co-registration approaches.
Atlas-ISTN: Joint Segmentation, Registration and Atlas Construction with Image-and-Spatial Transformer Networks
Dec 18, 2020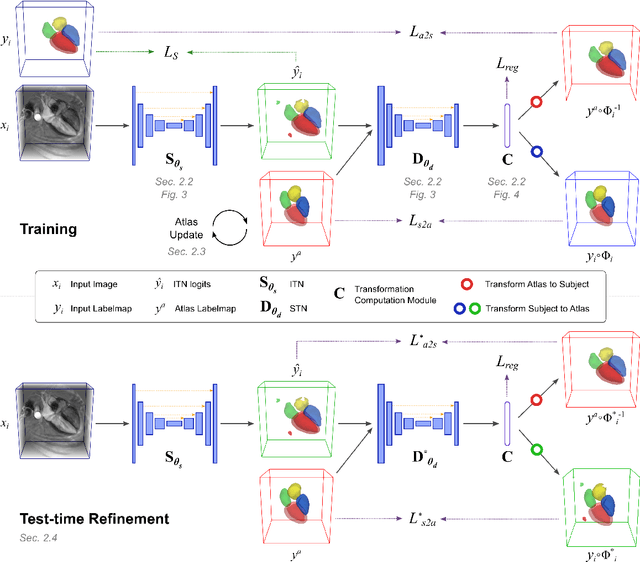
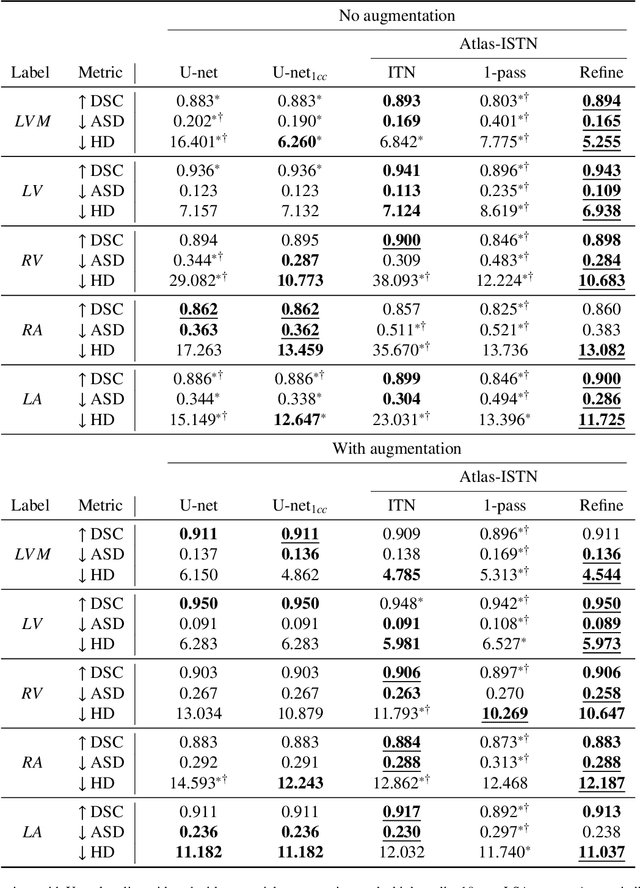
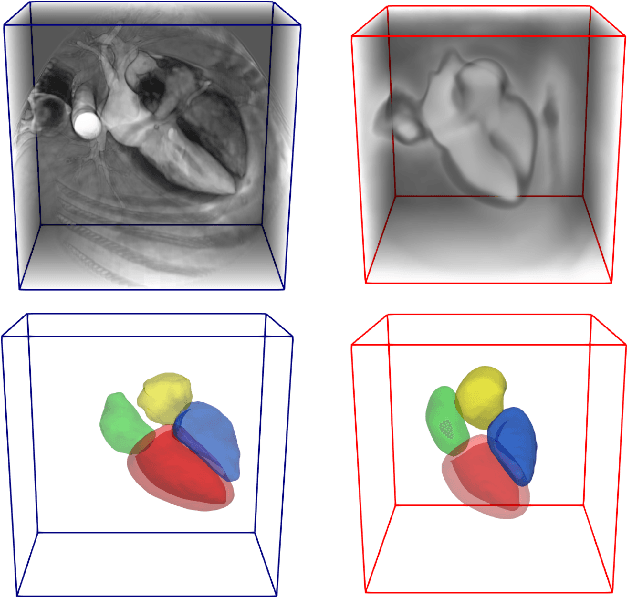
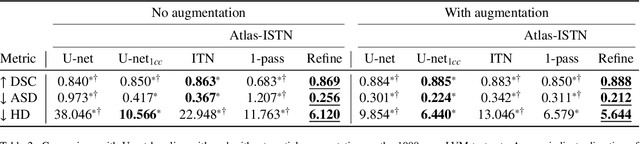
Abstract:Deep learning models for semantic segmentation are able to learn powerful representations for pixel-wise predictions, but are sensitive to noise at test time and do not guarantee a plausible topology. Image registration models on the other hand are able to warp known topologies to target images as a means of segmentation, but typically require large amounts of training data, and have not widely been benchmarked against pixel-wise segmentation models. We propose Atlas-ISTN, a framework that jointly learns segmentation and registration on 2D and 3D image data, and constructs a population-derived atlas in the process. Atlas-ISTN learns to segment multiple structures of interest and to register the constructed, topologically consistent atlas labelmap to an intermediate pixel-wise segmentation. Additionally, Atlas-ISTN allows for test time refinement of the model's parameters to optimize the alignment of the atlas labelmap to an intermediate pixel-wise segmentation. This process both mitigates for noise in the target image that can result in spurious pixel-wise predictions, as well as improves upon the one-pass prediction of the model. Benefits of the Atlas-ISTN framework are demonstrated qualitatively and quantitatively on 2D synthetic data and 3D cardiac computed tomography and brain magnetic resonance image data, out-performing both segmentation and registration baseline models. Atlas-ISTN also provides inter-subject correspondence of the structures of interest, enabling population-level shape and motion analysis.
Image-and-Spatial Transformer Networks for Structure-Guided Image Registration
Jul 22, 2019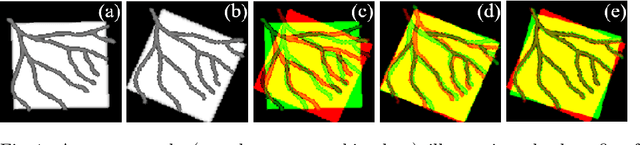
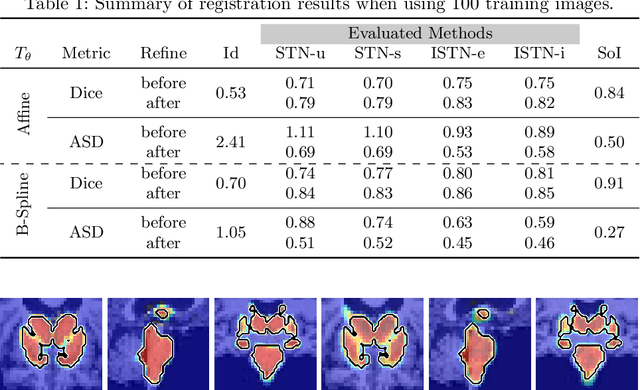
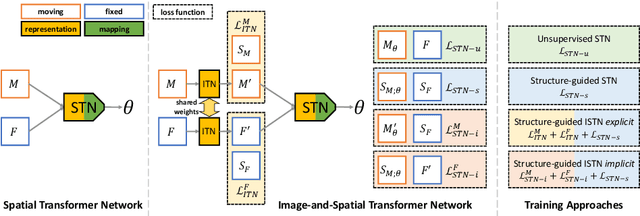
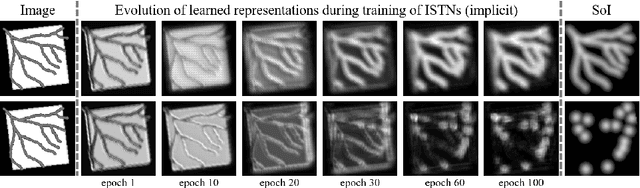
Abstract:Image registration with deep neural networks has become an active field of research and exciting avenue for a long standing problem in medical imaging. The goal is to learn a complex function that maps the appearance of input image pairs to parameters of a spatial transformation in order to align corresponding anatomical structures. We argue and show that the current direct, non-iterative approaches are sub-optimal, in particular if we seek accurate alignment of Structures-of-Interest (SoI). Information about SoI is often available at training time, for example, in form of segmentations or landmarks. We introduce a novel, generic framework, Image-and-Spatial Transformer Networks (ISTNs), to leverage SoI information allowing us to learn new image representations that are optimised for the downstream registration task. Thanks to these representations we can employ a test-specific, iterative refinement over the transformation parameters which yields highly accurate registration even with very limited training data. Performance is demonstrated on pairwise 3D brain registration and illustrative synthetic data.
Attention Gated Networks: Learning to Leverage Salient Regions in Medical Images
Aug 22, 2018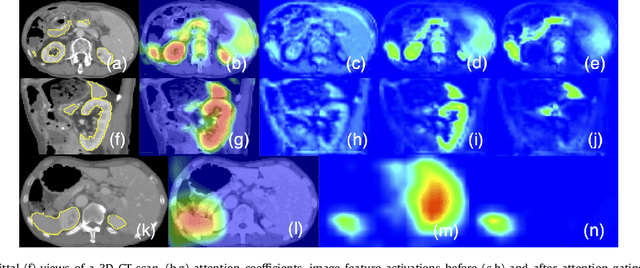
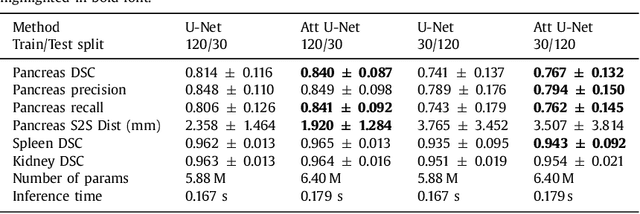
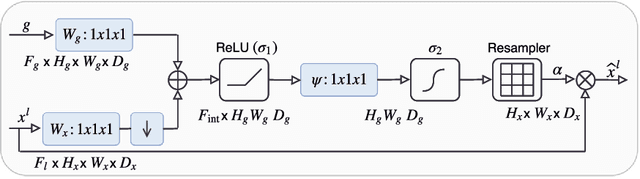
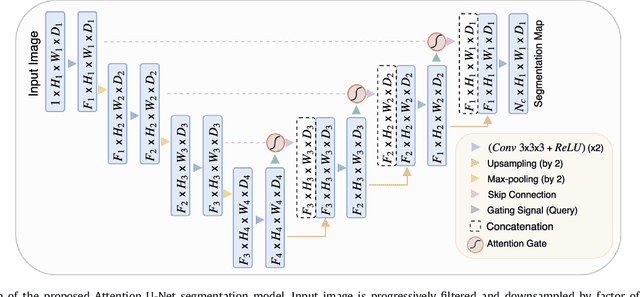
Abstract:We propose a novel attention gate (AG) model for medical image analysis that automatically learns to focus on target structures of varying shapes and sizes. Models trained with AGs implicitly learn to suppress irrelevant regions in an input image while highlighting salient features useful for a specific task. This enables us to eliminate the necessity of using explicit external tissue/organ localisation modules when using convolutional neural networks (CNNs). AGs can be easily integrated into standard CNN models such as VGG or U-Net architectures with minimal computational overhead while increasing the model sensitivity and prediction accuracy. The proposed AG models are evaluated on a variety of tasks, including medical image classification and segmentation. For classification, we demonstrate the use case of AGs in scan plane detection for fetal ultrasound screening. We show that the proposed attention mechanism can provide efficient object localisation while improving the overall prediction performance by reducing false positives. For segmentation, the proposed architecture is evaluated on two large 3D CT abdominal datasets with manual annotations for multiple organs. Experimental results show that AG models consistently improve the prediction performance of the base architectures across different datasets and training sizes while preserving computational efficiency. Moreover, AGs guide the model activations to be focused around salient regions, which provides better insights into how model predictions are made. The source code for the proposed AG models is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge