Julia A. Schnabel
Mask, Stitch, and Re-Sample: Enhancing Robustness and Generalizability in Anomaly Detection through Automatic Diffusion Models
May 31, 2023Abstract:The introduction of diffusion models in anomaly detection has paved the way for more effective and accurate image reconstruction in pathologies. However, the current limitations in controlling noise granularity hinder diffusion models' ability to generalize across diverse anomaly types and compromise the restoration of healthy tissues. To overcome these challenges, we propose AutoDDPM, a novel approach that enhances the robustness of diffusion models. AutoDDPM utilizes diffusion models to generate initial likelihood maps of potential anomalies and seamlessly integrates them with the original image. Through joint noised distribution re-sampling, AutoDDPM achieves harmonization and in-painting effects. Our study demonstrates the efficacy of AutoDDPM in replacing anomalous regions while preserving healthy tissues, considerably surpassing diffusion models' limitations. It also contributes valuable insights and analysis on the limitations of current diffusion models, promoting robust and interpretable anomaly detection in medical imaging - an essential aspect of building autonomous clinical decision systems with higher interpretability.
Deep Learning for Retrospective Motion Correction in MRI: A Comprehensive Review
May 11, 2023



Abstract:Motion represents one of the major challenges in magnetic resonance imaging (MRI). Since the MR signal is acquired in frequency space, any motion of the imaged object leads to complex artefacts in the reconstructed image in addition to other MR imaging artefacts. Deep learning has been frequently proposed for motion correction at several stages of the reconstruction process. The wide range of MR acquisition sequences, anatomies and pathologies of interest, and motion patterns (rigid vs. deformable and random vs. regular) makes a comprehensive solution unlikely. To facilitate the transfer of ideas between different applications, this review provides a detailed overview of proposed methods for learning-based motion correction in MRI together with their common challenges and potentials. This review identifies differences and synergies in underlying data usage, architectures and evaluation strategies. We critically discuss general trends and outline future directions, with the aim to enhance interaction between different application areas and research fields.
Deep Learning-Based Detection of Motion-Affected k-Space Lines for T2*-Weighted MRI
Mar 20, 2023Abstract:T2*-weighted gradient echo MR imaging is strongly impacted by subject head motion due to motion-related changes in B0 inhomogeneities. Within the oxygenation-sensitive mqBOLD protocol, even mild motion during the acquisition of the T2*-weighted data propagates into errors in derived quantitative parameter maps. In order to correct these images without the need of repeated measurements, we propose to learn a classification of motion-affected k-space lines. To test this, we perform realistic motion simulations including motion-induced field inhomogeneity changes for supervised training. To detect the presence of motion in each phase encoding line, we train a convolutional neural network, leveraging the multi-echo information of the T2*-weighted images. The proposed network accurately detects motion-affected k-space lines for simulated displacements of $\geq$ 0.5mm (accuracy on test set: 92.5%). Finally, we show example reconstructions where we include these classification labels as weights in the data consistency term of an iterative reconstruction procedure, opening up exciting opportunities of k-space line detection in combination with more powerful reconstruction methods.
3D Masked Autoencoders with Application to Anomaly Detection in Non-Contrast Enhanced Breast MRI
Mar 10, 2023Abstract:Self-supervised models allow (pre-)training on unlabeled data and therefore have the potential to overcome the need for large annotated cohorts. One leading self-supervised model is the masked autoencoder (MAE) which was developed on natural imaging data. The MAE is masking out a high fraction of visual transformer (ViT) input patches, to then recover the uncorrupted images as a pretraining task. In this work, we extend MAE to perform anomaly detection on breast magnetic resonance imaging (MRI). This new model, coined masked autoencoder for medical imaging (MAEMI) is trained on two non-contrast enhanced MRI sequences, aiming at lesion detection without the need for intravenous injection of contrast media and temporal image acquisition. During training, only non-cancerous images are presented to the model, with the purpose of localizing anomalous tumor regions during test time. We use a public dataset for model development. Performance of the architecture is evaluated in reference to subtraction images created from dynamic contrast enhanced (DCE)-MRI.
Fully transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study
Jan 23, 2023Abstract:Background: Deep learning (DL) can extract predictive and prognostic biomarkers from routine pathology slides in colorectal cancer. For example, a DL test for the diagnosis of microsatellite instability (MSI) in CRC has been approved in 2022. Current approaches rely on convolutional neural networks (CNNs). Transformer networks are outperforming CNNs and are replacing them in many applications, but have not been used for biomarker prediction in cancer at a large scale. In addition, most DL approaches have been trained on small patient cohorts, which limits their clinical utility. Methods: In this study, we developed a new fully transformer-based pipeline for end-to-end biomarker prediction from pathology slides. We combine a pre-trained transformer encoder and a transformer network for patch aggregation, capable of yielding single and multi-target prediction at patient level. We train our pipeline on over 9,000 patients from 10 colorectal cancer cohorts. Results: A fully transformer-based approach massively improves the performance, generalizability, data efficiency, and interpretability as compared with current state-of-the-art algorithms. After training on a large multicenter cohort, we achieve a sensitivity of 0.97 with a negative predictive value of 0.99 for MSI prediction on surgical resection specimens. We demonstrate for the first time that resection specimen-only training reaches clinical-grade performance on endoscopic biopsy tissue, solving a long-standing diagnostic problem. Interpretation: A fully transformer-based end-to-end pipeline trained on thousands of pathology slides yields clinical-grade performance for biomarker prediction on surgical resections and biopsies. Our new methods are freely available under an open source license.
Automated Quality Controlled Analysis of 2D Phase Contrast Cardiovascular Magnetic Resonance Imaging
Sep 28, 2022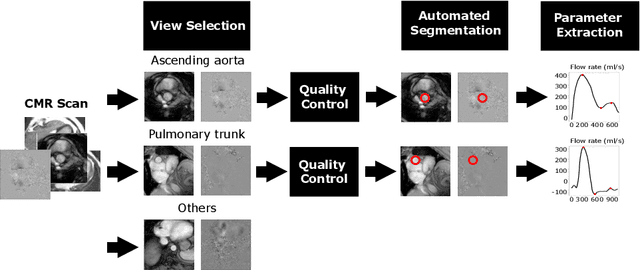
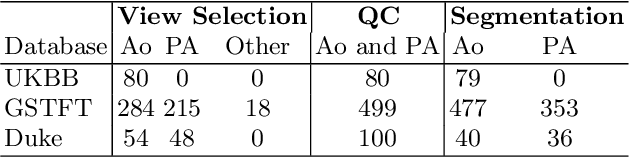
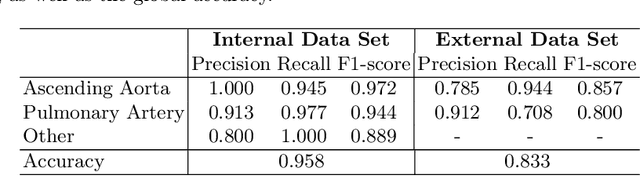
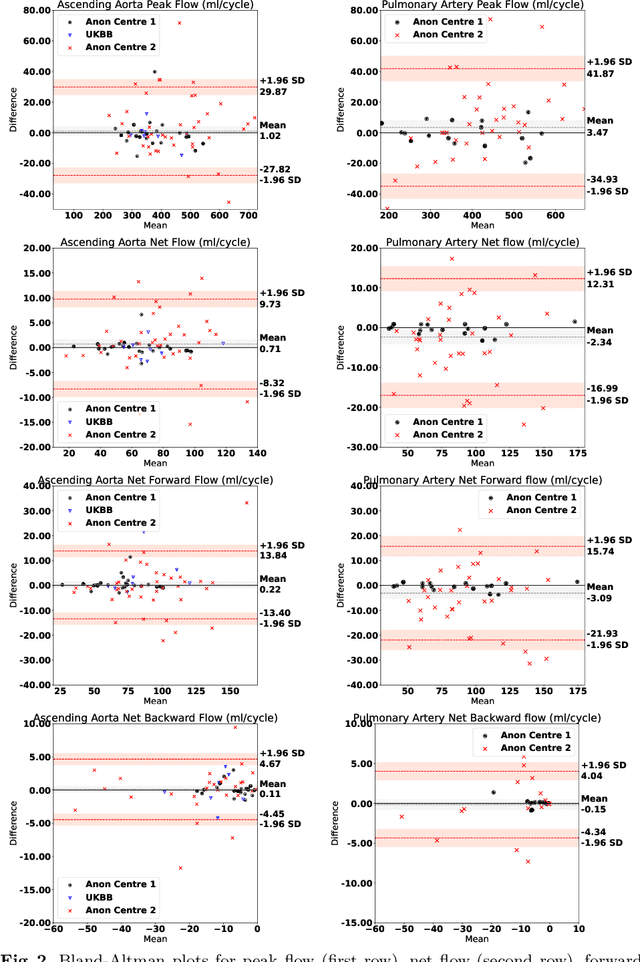
Abstract:Flow analysis carried out using phase contrast cardiac magnetic resonance imaging (PC-CMR) enables the quantification of important parameters that are used in the assessment of cardiovascular function. An essential part of this analysis is the identification of the correct CMR views and quality control (QC) to detect artefacts that could affect the flow quantification. We propose a novel deep learning based framework for the fully-automated analysis of flow from full CMR scans that first carries out these view selection and QC steps using two sequential convolutional neural networks, followed by automatic aorta and pulmonary artery segmentation to enable the quantification of key flow parameters. Accuracy values of 0.958 and 0.914 were obtained for view classification and QC, respectively. For segmentation, Dice scores were $>$0.969 and the Bland-Altman plots indicated excellent agreement between manual and automatic peak flow values. In addition, we tested our pipeline on an external validation data set, with results indicating good robustness of the pipeline. This work was carried out using multivendor clinical data consisting of 986 cases, indicating the potential for the use of this pipeline in a clinical setting.
Evaluation of 3D GANs for Lung Tissue Modelling in Pulmonary CT
Aug 17, 2022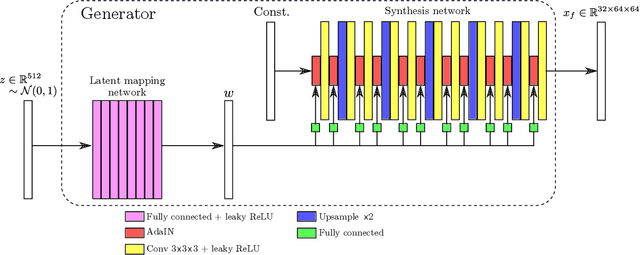
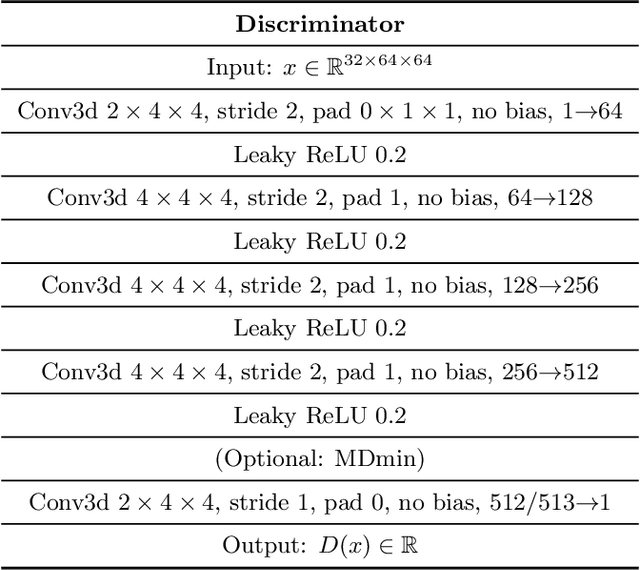
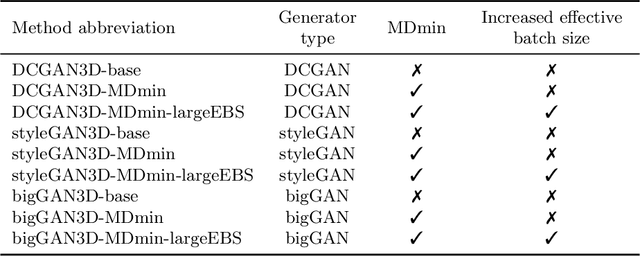

Abstract:GANs are able to model accurately the distribution of complex, high-dimensional datasets, e.g. images. This makes high-quality GANs useful for unsupervised anomaly detection in medical imaging. However, differences in training datasets such as output image dimensionality and appearance of semantically meaningful features mean that GAN models from the natural image domain may not work `out-of-the-box' for medical imaging, necessitating re-implementation and re-evaluation. In this work we adapt and evaluate three GAN models to the task of modelling 3D healthy image patches for pulmonary CT. To the best of our knowledge, this is the first time that such an evaluation has been performed. The DCGAN, styleGAN and the bigGAN architectures were investigated due to their ubiquity and high performance in natural image processing. We train different variants of these methods and assess their performance using the FID score. In addition, the quality of the generated images was evaluated by a human observer study, the ability of the networks to model 3D domain-specific features was investigated, and the structure of the GAN latent spaces was analysed. Results show that the 3D styleGAN produces realistic-looking images with meaningful 3D structure, but suffer from mode collapse which must be addressed during training to obtain samples diversity. Conversely, the 3D DCGAN models show a greater capacity for image variability, but at the cost of poor-quality images. The 3D bigGAN models provide an intermediate level of image quality, but most accurately model the distribution of selected semantically meaningful features. The results suggest that future development is required to realise a 3D GAN with sufficient capacity for patch-based lung CT anomaly detection and we offer recommendations for future areas of research, such as experimenting with other architectures and incorporation of position-encoding.
Placenta Segmentation in Ultrasound Imaging: Addressing Sources of Uncertainty and Limited Field-of-View
Jun 29, 2022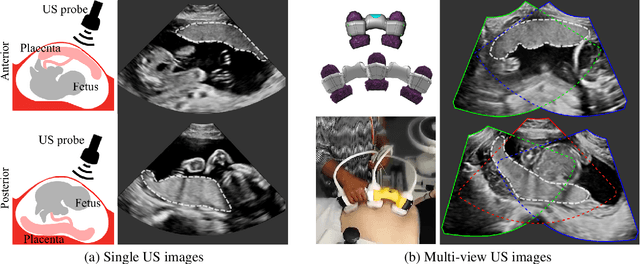
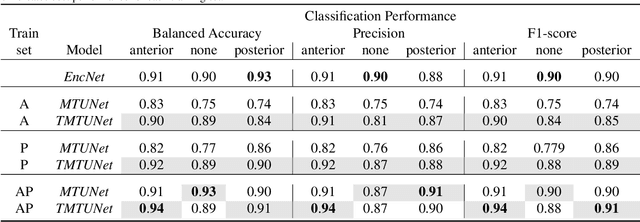
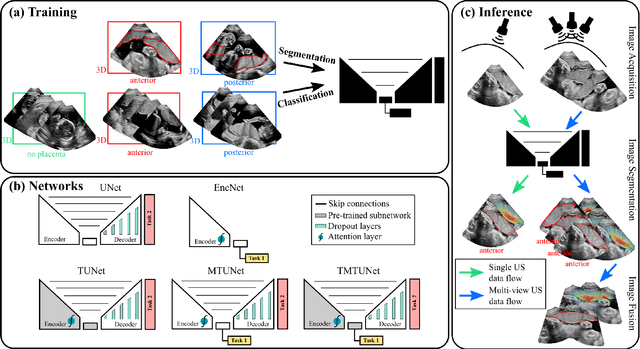
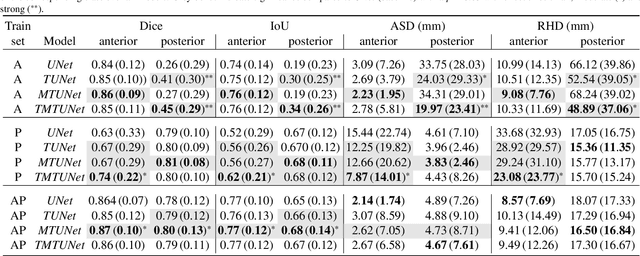
Abstract:Automatic segmentation of the placenta in fetal ultrasound (US) is challenging due to the (i) high diversity of placenta appearance, (ii) the restricted quality in US resulting in highly variable reference annotations, and (iii) the limited field-of-view of US prohibiting whole placenta assessment at late gestation. In this work, we address these three challenges with a multi-task learning approach that combines the classification of placental location (e.g., anterior, posterior) and semantic placenta segmentation in a single convolutional neural network. Through the classification task the model can learn from larger and more diverse datasets while improving the accuracy of the segmentation task in particular in limited training set conditions. With this approach we investigate the variability in annotations from multiple raters and show that our automatic segmentations (Dice of 0.86 for anterior and 0.83 for posterior placentas) achieve human-level performance as compared to intra- and inter-observer variability. Lastly, our approach can deliver whole placenta segmentation using a multi-view US acquisition pipeline consisting of three stages: multi-probe image acquisition, image fusion and image segmentation. This results in high quality segmentation of larger structures such as the placenta in US with reduced image artifacts which are beyond the field-of-view of single probes.
What do we learn? Debunking the Myth of Unsupervised Outlier Detection
Jun 08, 2022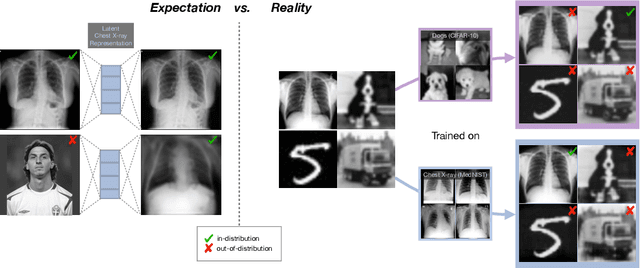
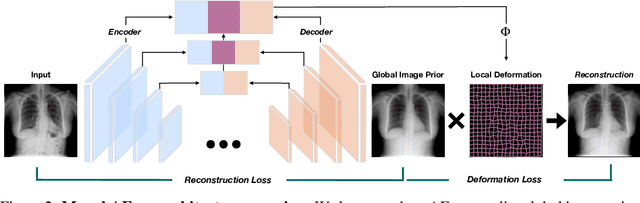
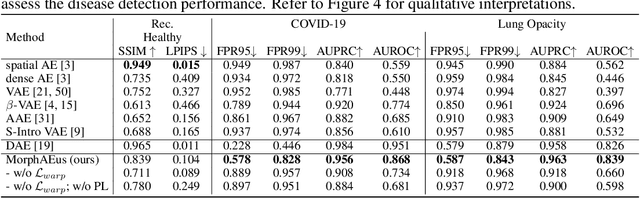
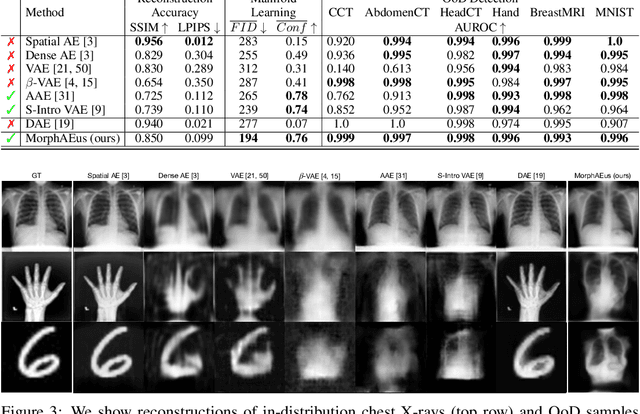
Abstract:Even though auto-encoders (AEs) have the desirable property of learning compact representations without labels and have been widely applied to out-of-distribution (OoD) detection, they are generally still poorly understood and are used incorrectly in detecting outliers where the normal and abnormal distributions are strongly overlapping. In general, the learned manifold is assumed to contain key information that is only important for describing samples within the training distribution, and that the reconstruction of outliers leads to high residual errors. However, recent work suggests that AEs are likely to be even better at reconstructing some types of OoD samples. In this work, we challenge this assumption and investigate what auto-encoders actually learn when they are posed to solve two different tasks. First, we propose two metrics based on the Fr\'echet inception distance (FID) and confidence scores of a trained classifier to assess whether AEs can learn the training distribution and reliably recognize samples from other domains. Second, we investigate whether AEs are able to synthesize normal images from samples with abnormal regions, on a more challenging lung pathology detection task. We have found that state-of-the-art (SOTA) AEs are either unable to constrain the latent manifold and allow reconstruction of abnormal patterns, or they are failing to accurately restore the inputs from their latent distribution, resulting in blurred or misaligned reconstructions. We propose novel deformable auto-encoders (MorphAEus) to learn perceptually aware global image priors and locally adapt their morphometry based on estimated dense deformation fields. We demonstrate superior performance over unsupervised methods in detecting OoD and pathology.
A Deep Learning-based Integrated Framework for Quality-aware Undersampled Cine Cardiac MRI Reconstruction and Analysis
May 02, 2022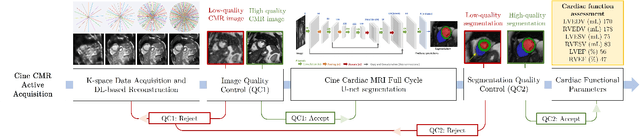
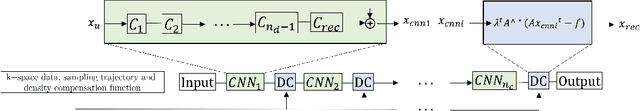
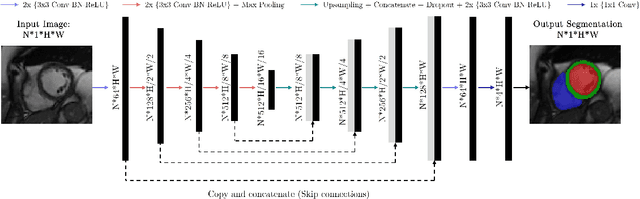
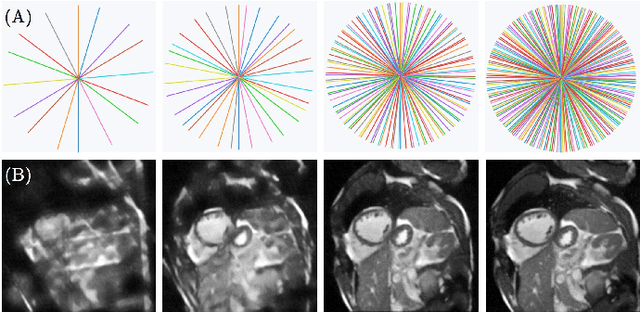
Abstract:Cine cardiac magnetic resonance (CMR) imaging is considered the gold standard for cardiac function evaluation. However, cine CMR acquisition is inherently slow and in recent decades considerable effort has been put into accelerating scan times without compromising image quality or the accuracy of derived results. In this paper, we present a fully-automated, quality-controlled integrated framework for reconstruction, segmentation and downstream analysis of undersampled cine CMR data. The framework enables active acquisition of radial k-space data, in which acquisition can be stopped as soon as acquired data are sufficient to produce high quality reconstructions and segmentations. This results in reduced scan times and automated analysis, enabling robust and accurate estimation of functional biomarkers. To demonstrate the feasibility of the proposed approach, we perform realistic simulations of radial k-space acquisitions on a dataset of subjects from the UK Biobank and present results on in-vivo cine CMR k-space data collected from healthy subjects. The results demonstrate that our method can produce quality-controlled images in a mean scan time reduced from 12 to 4 seconds per slice, and that image quality is sufficient to allow clinically relevant parameters to be automatically estimated to within 5% mean absolute difference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge