Arjun Nair
A 3D deep learning classifier and its explainability when assessing coronary artery disease
Jul 29, 2023Abstract:Early detection and diagnosis of coronary artery disease (CAD) could save lives and reduce healthcare costs. In this study, we propose a 3D Resnet-50 deep learning model to directly classify normal subjects and CAD patients on computed tomography coronary angiography images. Our proposed method outperforms a 2D Resnet-50 model by 23.65%. Explainability is also provided by using a Grad-GAM. Furthermore, we link the 3D CAD classification to a 2D two-class semantic segmentation for improved explainability and accurate abnormality localisation.
STRUDEL: Structured Dialogue Summarization for Dialogue Comprehension
Dec 24, 2022



Abstract:Abstractive dialogue summarization has long been viewed as an important standalone task in natural language processing, but no previous work has explored the possibility of whether abstractive dialogue summarization can also be used as a means to boost an NLP system's performance on other important dialogue comprehension tasks. In this paper, we propose a novel type of dialogue summarization task - STRUctured DiaLoguE Summarization - that can help pre-trained language models to better understand dialogues and improve their performance on important dialogue comprehension tasks. We further collect human annotations of STRUDEL summaries over 400 dialogues and introduce a new STRUDEL dialogue comprehension modeling framework that integrates STRUDEL into a graph-neural-network-based dialogue reasoning module over transformer encoder language models to improve their dialogue comprehension abilities. In our empirical experiments on two important downstream dialogue comprehension tasks - dialogue question answering and dialogue response prediction - we show that our STRUDEL dialogue comprehension model can significantly improve the dialogue comprehension performance of transformer encoder language models.
Evaluation of 3D GANs for Lung Tissue Modelling in Pulmonary CT
Aug 17, 2022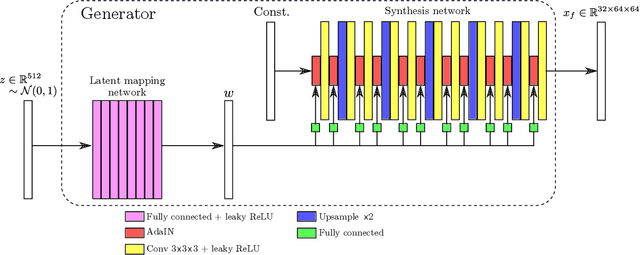
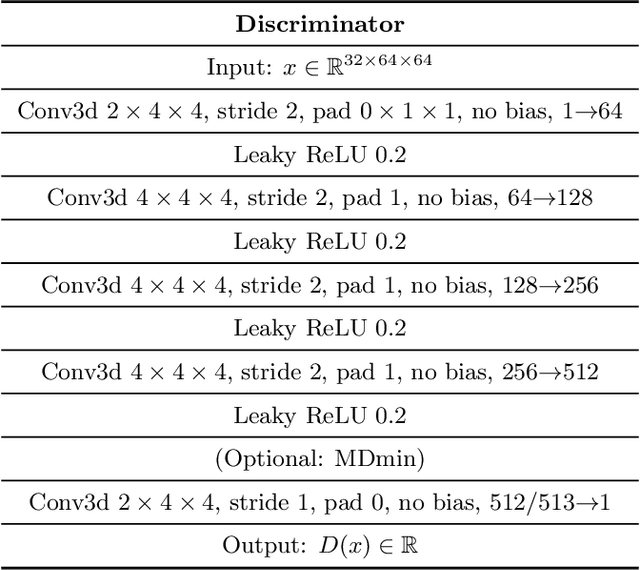
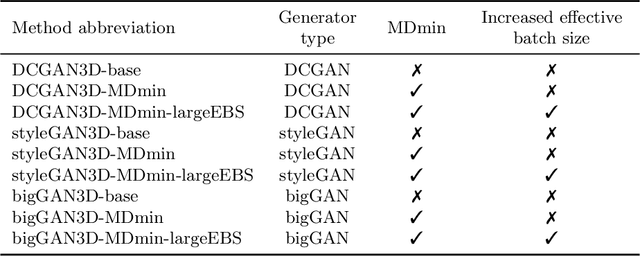

Abstract:GANs are able to model accurately the distribution of complex, high-dimensional datasets, e.g. images. This makes high-quality GANs useful for unsupervised anomaly detection in medical imaging. However, differences in training datasets such as output image dimensionality and appearance of semantically meaningful features mean that GAN models from the natural image domain may not work `out-of-the-box' for medical imaging, necessitating re-implementation and re-evaluation. In this work we adapt and evaluate three GAN models to the task of modelling 3D healthy image patches for pulmonary CT. To the best of our knowledge, this is the first time that such an evaluation has been performed. The DCGAN, styleGAN and the bigGAN architectures were investigated due to their ubiquity and high performance in natural image processing. We train different variants of these methods and assess their performance using the FID score. In addition, the quality of the generated images was evaluated by a human observer study, the ability of the networks to model 3D domain-specific features was investigated, and the structure of the GAN latent spaces was analysed. Results show that the 3D styleGAN produces realistic-looking images with meaningful 3D structure, but suffer from mode collapse which must be addressed during training to obtain samples diversity. Conversely, the 3D DCGAN models show a greater capacity for image variability, but at the cost of poor-quality images. The 3D bigGAN models provide an intermediate level of image quality, but most accurately model the distribution of selected semantically meaningful features. The results suggest that future development is required to realise a 3D GAN with sufficient capacity for patch-based lung CT anomaly detection and we offer recommendations for future areas of research, such as experimenting with other architectures and incorporation of position-encoding.
Enhancing Cancer Prediction in Challenging Screen-Detected Incident Lung Nodules Using Time-Series Deep Learning
Mar 30, 2022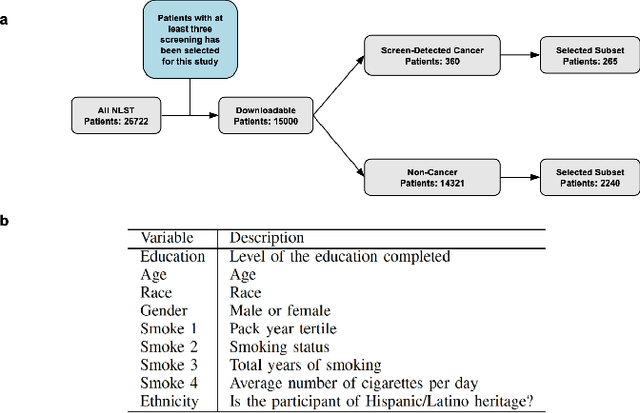
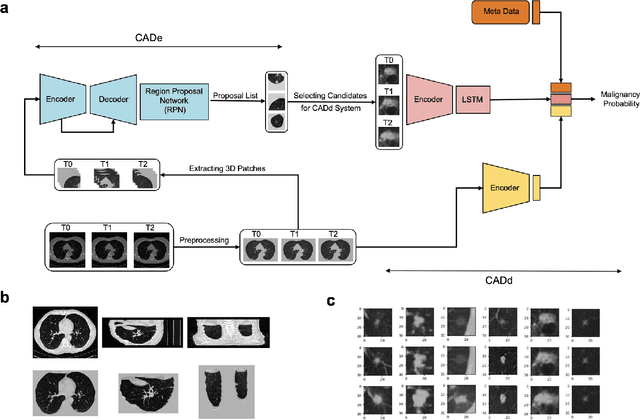
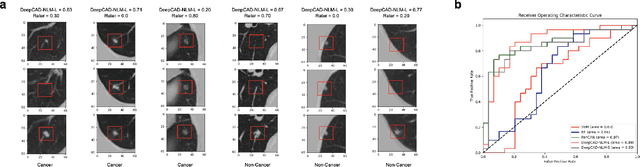
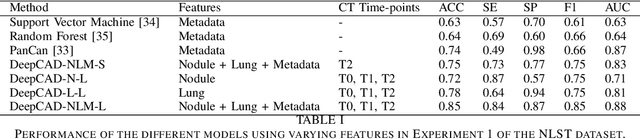
Abstract:Lung cancer is the leading cause of cancer-related mortality worldwide. Lung cancer screening (LCS) using annual low-dose computed tomography (CT) scanning has been proven to significantly reduce lung cancer mortality by detecting cancerous lung nodules at an earlier stage. Improving risk stratification of malignancy risk in lung nodules can be enhanced using machine/deep learning algorithms. However most existing algorithms: a) have primarily assessed single time-point CT data alone thereby failing to utilize the inherent advantages contained within longitudinal imaging datasets; b) have not integrated into computer models pertinent clinical data that might inform risk prediction; c) have not assessed algorithm performance on the spectrum of nodules that are most challenging for radiologists to interpret and where assistance from analytic tools would be most beneficial. Here we show the performance of our time-series deep learning model (DeepCAD-NLM-L) which integrates multi-model information across three longitudinal data domains: nodule-specific, lung-specific, and clinical demographic data. We compared our time-series deep learning model to a) radiologist performance on CTs from the National Lung Screening Trial enriched with the most challenging nodules for diagnosis; b) a nodule management algorithm from a North London LCS study (SUMMIT). Our model demonstrated comparable and complementary performance to radiologists when interpreting challenging lung nodules and showed improved performance (AUC=88\%) against models utilizing single time-point data only. The results emphasise the importance of time-series, multi-modal analysis when interpreting malignancy risk in LCS.
CONFIT: Toward Faithful Dialogue Summarization with Linguistically-Informed Contrastive Fine-tuning
Dec 16, 2021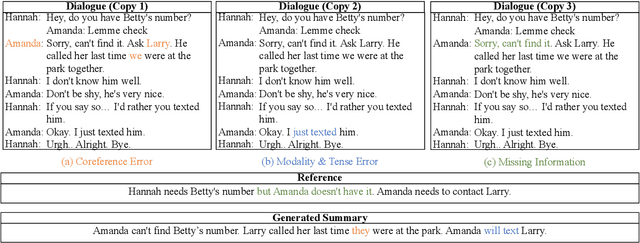
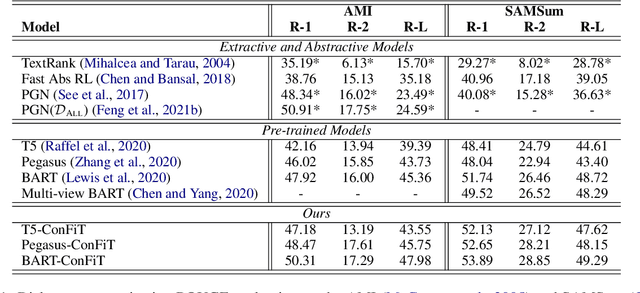
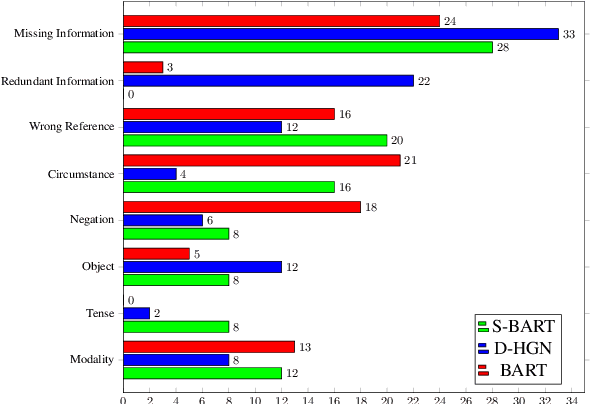
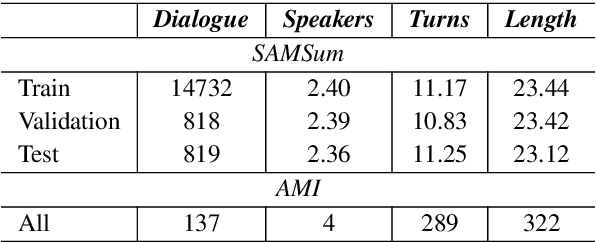
Abstract:Factual inconsistencies in generated summaries severely limit the practical applications of abstractive dialogue summarization. Although significant progress has been achieved by using pre-trained models, substantial amounts of hallucinated content are found during the human evaluation. Pre-trained models are most commonly fine-tuned with cross-entropy loss for text summarization, which may not be an optimal strategy. In this work, we provide a typology of factual errors with annotation data to highlight the types of errors and move away from a binary understanding of factuality. We further propose a training strategy that improves the factual consistency and overall quality of summaries via a novel contrastive fine-tuning, called ConFiT. Based on our linguistically-informed typology of errors, we design different modular objectives that each target a specific type. Specifically, we utilize hard negative samples with errors to reduce the generation of factual inconsistency. In order to capture the key information between speakers, we also design a dialogue-specific loss. Using human evaluation and automatic faithfulness metrics, we show that our model significantly reduces all kinds of factual errors on the dialogue summarization, SAMSum corpus. Moreover, our model could be generalized to the meeting summarization, AMI corpus, and it produces significantly higher scores than most of the baselines on both datasets regarding word-overlap metrics.
Is MC Dropout Bayesian?
Oct 08, 2021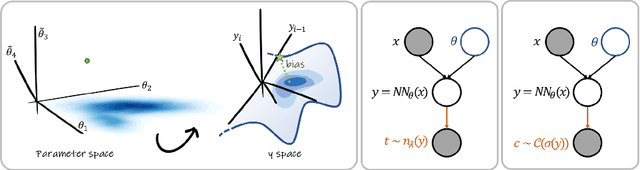
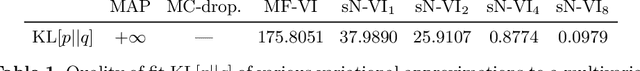
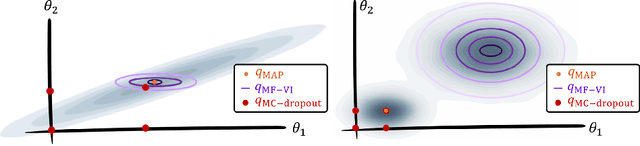

Abstract:MC Dropout is a mainstream "free lunch" method in medical imaging for approximate Bayesian computations (ABC). Its appeal is to solve out-of-the-box the daunting task of ABC and uncertainty quantification in Neural Networks (NNs); to fall within the variational inference (VI) framework; and to propose a highly multimodal, faithful predictive posterior. We question the properties of MC Dropout for approximate inference, as in fact MC Dropout changes the Bayesian model; its predictive posterior assigns $0$ probability to the true model on closed-form benchmarks; the multimodality of its predictive posterior is not a property of the true predictive posterior but a design artefact. To address the need for VI on arbitrary models, we share a generic VI engine within the pytorch framework. The code includes a carefully designed implementation of structured (diagonal plus low-rank) multivariate normal variational families, and mixtures thereof. It is intended as a go-to no-free-lunch approach, addressing shortcomings of mean-field VI with an adjustable trade-off between expressivity and computational complexity.
The Pitfalls of Sample Selection: A Case Study on Lung Nodule Classification
Aug 11, 2021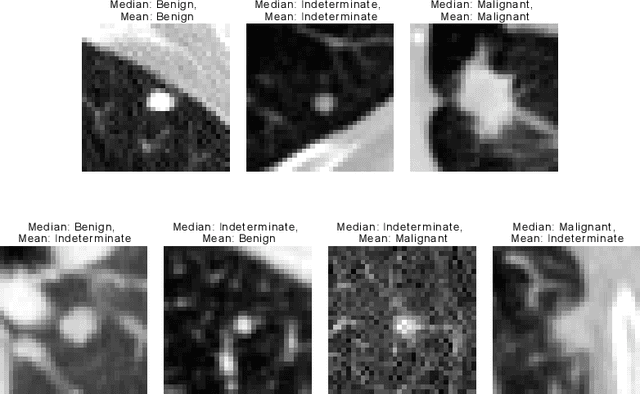
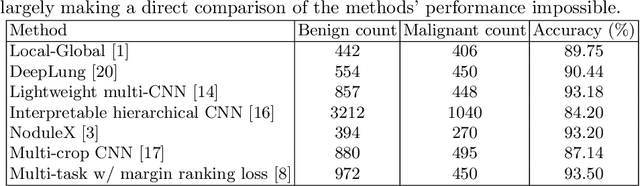
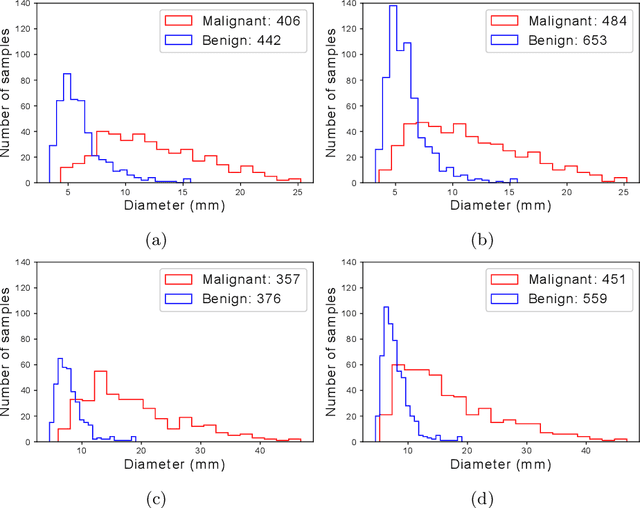
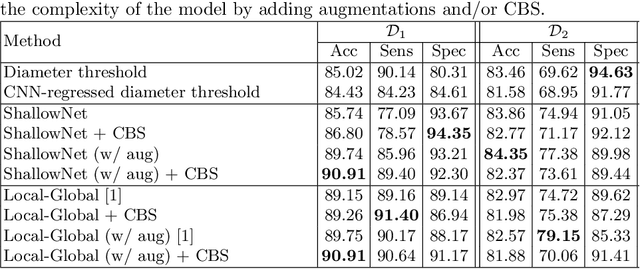
Abstract:Using publicly available data to determine the performance of methodological contributions is important as it facilitates reproducibility and allows scrutiny of the published results. In lung nodule classification, for example, many works report results on the publicly available LIDC dataset. In theory, this should allow a direct comparison of the performance of proposed methods and assess the impact of individual contributions. When analyzing seven recent works, however, we find that each employs a different data selection process, leading to largely varying total number of samples and ratios between benign and malignant cases. As each subset will have different characteristics with varying difficulty for classification, a direct comparison between the proposed methods is thus not always possible, nor fair. We study the particular effect of truthing when aggregating labels from multiple experts. We show that specific choices can have severe impact on the data distribution where it may be possible to achieve superior performance on one sample distribution but not on another. While we show that we can further improve on the state-of-the-art on one sample selection, we also find that on a more challenging sample selection, on the same database, the more advanced models underperform with respect to very simple baseline methods, highlighting that the selected data distribution may play an even more important role than the model architecture. This raises concerns about the validity of claimed methodological contributions. We believe the community should be aware of these pitfalls and make recommendations on how these can be avoided in future work.
The Effect of the Loss on Generalization: Empirical Study on Synthetic Lung Nodule Data
Aug 10, 2021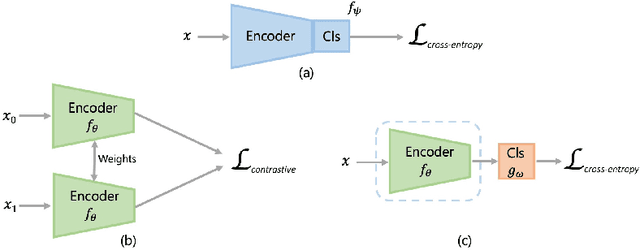
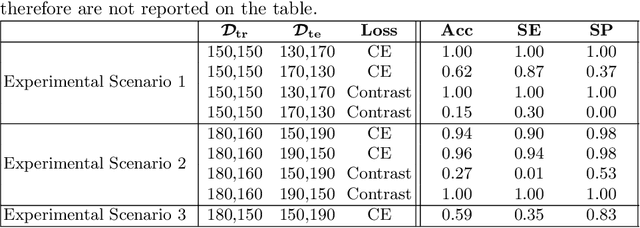
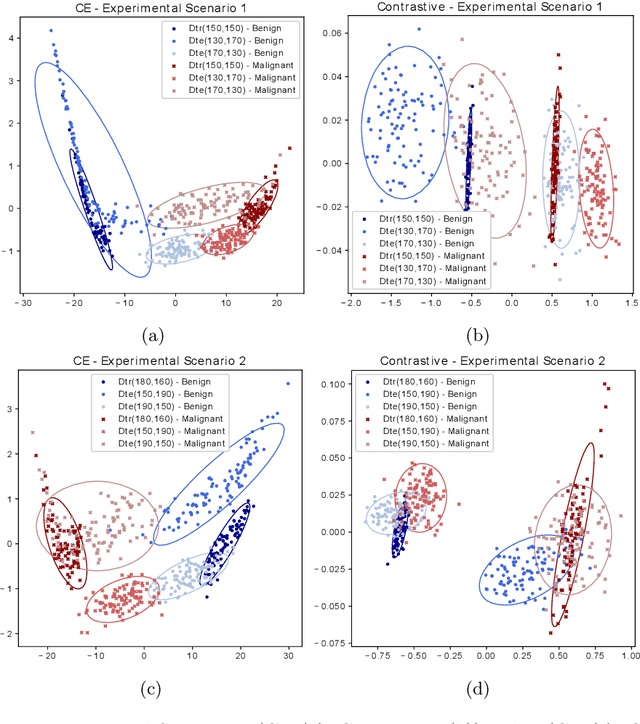
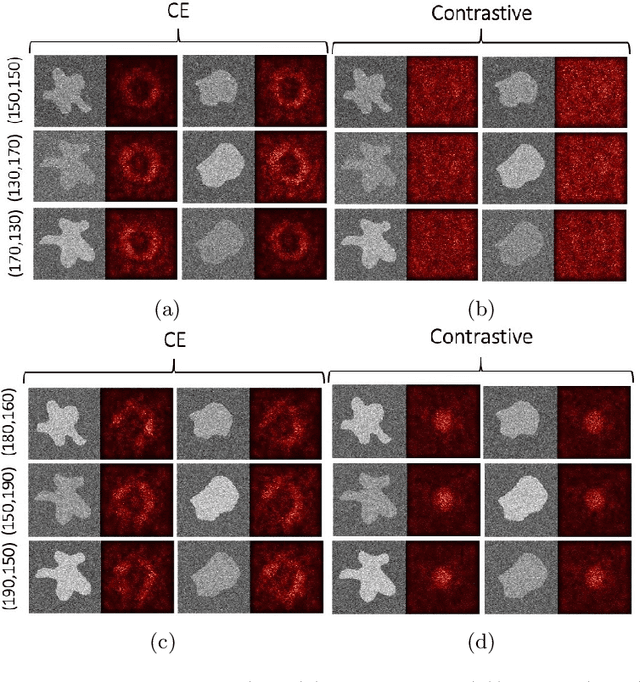
Abstract:Convolutional Neural Networks (CNNs) are widely used for image classification in a variety of fields, including medical imaging. While most studies deploy cross-entropy as the loss function in such tasks, a growing number of approaches have turned to a family of contrastive learning-based losses. Even though performance metrics such as accuracy, sensitivity and specificity are regularly used for the evaluation of CNN classifiers, the features that these classifiers actually learn are rarely identified and their effect on the classification performance on out-of-distribution test samples is insufficiently explored. In this paper, motivated by the real-world task of lung nodule classification, we investigate the features that a CNN learns when trained and tested on different distributions of a synthetic dataset with controlled modes of variation. We show that different loss functions lead to different features being learned and consequently affect the generalization ability of the classifier on unseen data. This study provides some important insights into the design of deep learning solutions for medical imaging tasks.
Bayesian analysis of the prevalence bias: learning and predicting from imbalanced data
Jul 31, 2021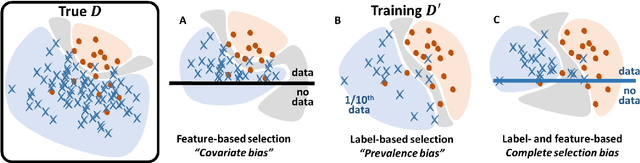
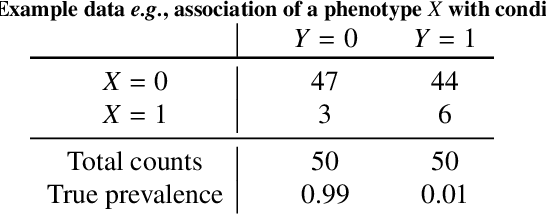
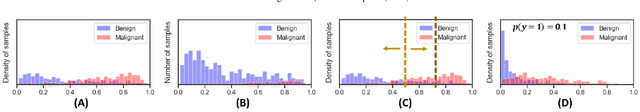
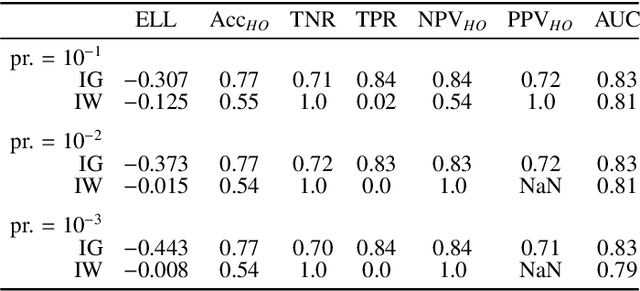
Abstract:Datasets are rarely a realistic approximation of the target population. Say, prevalence is misrepresented, image quality is above clinical standards, etc. This mismatch is known as sampling bias. Sampling biases are a major hindrance for machine learning models. They cause significant gaps between model performance in the lab and in the real world. Our work is a solution to prevalence bias. Prevalence bias is the discrepancy between the prevalence of a pathology and its sampling rate in the training dataset, introduced upon collecting data or due to the practioner rebalancing the training batches. This paper lays the theoretical and computational framework for training models, and for prediction, in the presence of prevalence bias. Concretely a bias-corrected loss function, as well as bias-corrected predictive rules, are derived under the principles of Bayesian risk minimization. The loss exhibits a direct connection to the information gain. It offers a principled alternative to heuristic training losses and complements test-time procedures based on selecting an operating point from summary curves. It integrates seamlessly in the current paradigm of (deep) learning using stochastic backpropagation and naturally with Bayesian models.
Modelling Airway Geometry as Stock Market Data using Bayesian Changepoint Detection
Jun 28, 2019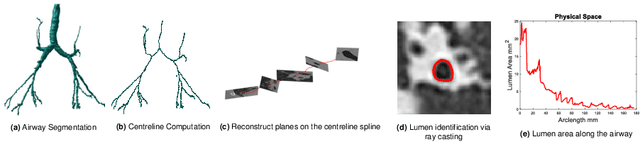
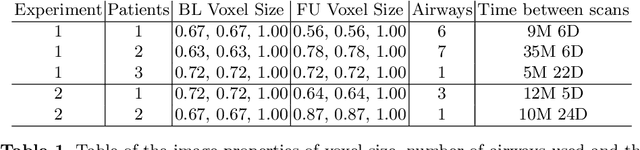
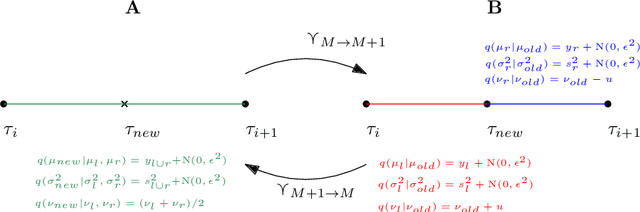
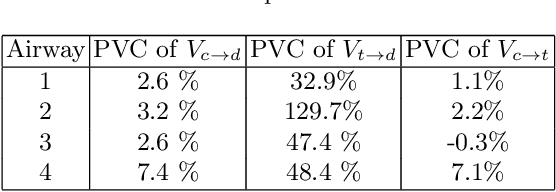
Abstract:Numerous lung diseases, such as idiopathic pulmonary fibrosis (IPF), exhibit dilation of the airways. Accurate measurement of dilatation enables assessment of the progression of disease. Unfortunately the combination of image noise and airway bifurcations causes high variability in the profiles of cross-sectional areas, rendering the identification of affected regions very difficult. Here we introduce a noise-robust method for automatically detecting the location of progressive airway dilatation given two profiles of the same airway acquired at different time points. We propose a probabilistic model of abrupt relative variations between profiles and perform inference via Reversible Jump Markov Chain Monte Carlo sampling. We demonstrate the efficacy of the proposed method on two datasets; (i) images of healthy airways with simulated dilatation; (ii) pairs of real images of IPF-affected airways acquired at 1 year intervals. Our model is able to detect the starting location of airway dilatation with an accuracy of 2.5mm on simulated data. The experiments on the IPF dataset display reasonable agreement with radiologists. We can compute a relative change in airway volume that may be useful for quantifying IPF disease progression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge