Jiliu Zhou
Triplet-constraint Transformer with Multi-scale Refinement for Dose Prediction in Radiotherapy
Feb 07, 2024



Abstract:Radiotherapy is a primary treatment for cancers with the aim of applying sufficient radiation dose to the planning target volume (PTV) while minimizing dose hazards to the organs at risk (OARs). Convolutional neural networks (CNNs) have automated the radiotherapy plan-making by predicting the dose maps. However, current CNN-based methods ignore the remarkable dose difference in the dose map, i.e., high dose value in the interior PTV while low value in the exterior PTV, leading to a suboptimal prediction. In this paper, we propose a triplet-constraint transformer (TCtrans) with multi-scale refinement to predict the high-quality dose distribution. Concretely, a novel PTV-guided triplet constraint is designed to refine dose feature representations in the interior and exterior PTV by utilizing the explicit geometry of PTV. Furthermore, we introduce a multi-scale refinement (MSR) module to effectively fulfill the triplet constraint in different decoding layers with multiple scales. Besides, a transformer encoder is devised to learn the important global dosimetric knowledge. Experiments on a clinical cervical cancer dataset demonstrate the superiority of our method.
Image2Points:A 3D Point-based Context Clusters GAN for High-Quality PET Image Reconstruction
Feb 01, 2024Abstract:To obtain high-quality Positron emission tomography (PET) images while minimizing radiation exposure, numerous methods have been proposed to reconstruct standard-dose PET (SPET) images from the corresponding low-dose PET (LPET) images. However, these methods heavily rely on voxel-based representations, which fall short of adequately accounting for the precise structure and fine-grained context, leading to compromised reconstruction. In this paper, we propose a 3D point-based context clusters GAN, namely PCC-GAN, to reconstruct high-quality SPET images from LPET. Specifically, inspired by the geometric representation power of points, we resort to a point-based representation to enhance the explicit expression of the image structure, thus facilitating the reconstruction with finer details. Moreover, a context clustering strategy is applied to explore the contextual relationships among points, which mitigates the ambiguities of small structures in the reconstructed images. Experiments on both clinical and phantom datasets demonstrate that our PCC-GAN outperforms the state-of-the-art reconstruction methods qualitatively and quantitatively. Code is available at https://github.com/gluucose/PCCGAN.
Diffusion-based Radiotherapy Dose Prediction Guided by Inter-slice Aware Structure Encoding
Nov 06, 2023



Abstract:Deep learning (DL) has successfully automated dose distribution prediction in radiotherapy planning, enhancing both efficiency and quality. However, existing methods suffer from the over-smoothing problem for their commonly used L1 or L2 loss with posterior average calculations. To alleviate this limitation, we propose a diffusion model-based method (DiffDose) for predicting the radiotherapy dose distribution of cancer patients. Specifically, the DiffDose model contains a forward process and a reverse process. In the forward process, DiffDose transforms dose distribution maps into pure Gaussian noise by gradually adding small noise and a noise predictor is simultaneously trained to estimate the noise added at each timestep. In the reverse process, it removes the noise from the pure Gaussian noise in multiple steps with the well-trained noise predictor and finally outputs the predicted dose distribution maps...
Contrastive Diffusion Model with Auxiliary Guidance for Coarse-to-Fine PET Reconstruction
Aug 20, 2023Abstract:To obtain high-quality positron emission tomography (PET) scans while reducing radiation exposure to the human body, various approaches have been proposed to reconstruct standard-dose PET (SPET) images from low-dose PET (LPET) images. One widely adopted technique is the generative adversarial networks (GANs), yet recently, diffusion probabilistic models (DPMs) have emerged as a compelling alternative due to their improved sample quality and higher log-likelihood scores compared to GANs. Despite this, DPMs suffer from two major drawbacks in real clinical settings, i.e., the computationally expensive sampling process and the insufficient preservation of correspondence between the conditioning LPET image and the reconstructed PET (RPET) image. To address the above limitations, this paper presents a coarse-to-fine PET reconstruction framework that consists of a coarse prediction module (CPM) and an iterative refinement module (IRM). The CPM generates a coarse PET image via a deterministic process, and the IRM samples the residual iteratively. By delegating most of the computational overhead to the CPM, the overall sampling speed of our method can be significantly improved. Furthermore, two additional strategies, i.e., an auxiliary guidance strategy and a contrastive diffusion strategy, are proposed and integrated into the reconstruction process, which can enhance the correspondence between the LPET image and the RPET image, further improving clinical reliability. Extensive experiments on two human brain PET datasets demonstrate that our method outperforms the state-of-the-art PET reconstruction methods. The source code is available at \url{https://github.com/Show-han/PET-Reconstruction}.
TriDo-Former: A Triple-Domain Transformer for Direct PET Reconstruction from Low-Dose Sinograms
Aug 10, 2023Abstract:To obtain high-quality positron emission tomography (PET) images while minimizing radiation exposure, various methods have been proposed for reconstructing standard-dose PET (SPET) images from low-dose PET (LPET) sinograms directly. However, current methods often neglect boundaries during sinogram-to-image reconstruction, resulting in high-frequency distortion in the frequency domain and diminished or fuzzy edges in the reconstructed images. Furthermore, the convolutional architectures, which are commonly used, lack the ability to model long-range non-local interactions, potentially leading to inaccurate representations of global structures. To alleviate these problems, we propose a transformer-based model that unites triple domains of sinogram, image, and frequency for direct PET reconstruction, namely TriDo-Former. Specifically, the TriDo-Former consists of two cascaded networks, i.e., a sinogram enhancement transformer (SE-Former) for denoising the input LPET sinograms and a spatial-spectral reconstruction transformer (SSR-Former) for reconstructing SPET images from the denoised sinograms. Different from the vanilla transformer that splits an image into 2D patches, based specifically on the PET imaging mechanism, our SE-Former divides the sinogram into 1D projection view angles to maintain its inner-structure while denoising, preventing the noise in the sinogram from prorogating into the image domain. Moreover, to mitigate high-frequency distortion and improve reconstruction details, we integrate global frequency parsers (GFPs) into SSR-Former. The GFP serves as a learnable frequency filter that globally adjusts the frequency components in the frequency domain, enforcing the network to restore high-frequency details resembling real SPET images. Validations on a clinical dataset demonstrate that our TriDo-Former outperforms the state-of-the-art methods qualitatively and quantitatively.
DiffDP: Radiotherapy Dose Prediction via a Diffusion Model
Jul 19, 2023



Abstract:Currently, deep learning (DL) has achieved the automatic prediction of dose distribution in radiotherapy planning, enhancing its efficiency and quality. However, existing methods suffer from the over-smoothing problem for their commonly used L_1 or L_2 loss with posterior average calculations. To alleviate this limitation, we innovatively introduce a diffusion-based dose prediction (DiffDP) model for predicting the radiotherapy dose distribution of cancer patients. Specifically, the DiffDP model contains a forward process and a reverse process. In the forward process, DiffDP gradually transforms dose distribution maps into Gaussian noise by adding small noise and trains a noise predictor to predict the noise added in each timestep. In the reverse process, it removes the noise from the original Gaussian noise in multiple steps with the well-trained noise predictor and finally outputs the predicted dose distribution map. To ensure the accuracy of the prediction, we further design a structure encoder to extract anatomical information from patient anatomy images and enable the noise predictor to be aware of the dose constraints within several essential organs, i.e., the planning target volume and organs at risk. Extensive experiments on an in-house dataset with 130 rectum cancer patients demonstrate the s
LDP-Net: An Unsupervised Pansharpening Network Based on Learnable Degradation Processes
Nov 24, 2021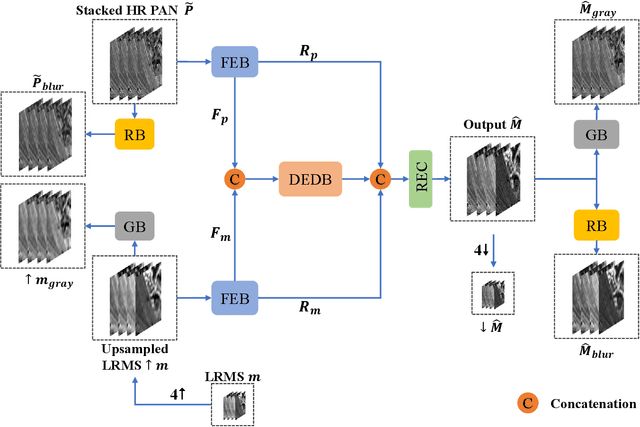
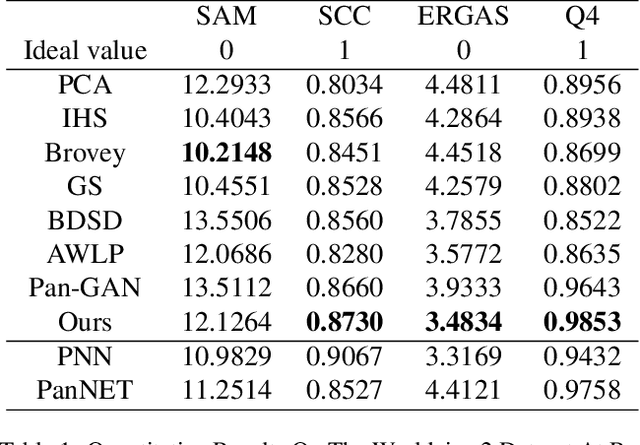
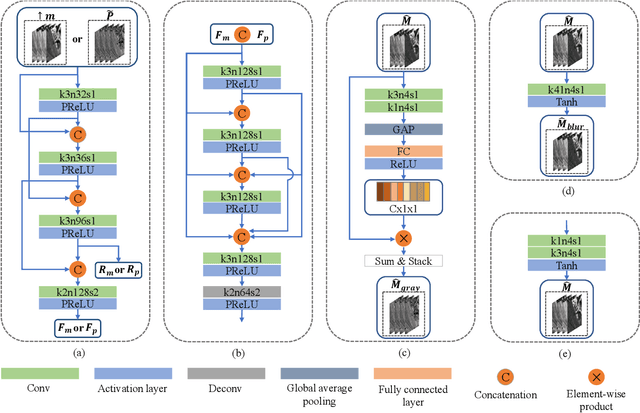
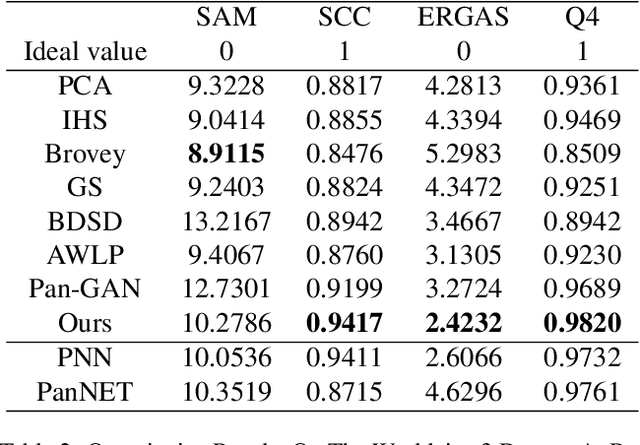
Abstract:Pansharpening in remote sensing image aims at acquiring a high-resolution multispectral (HRMS) image directly by fusing a low-resolution multispectral (LRMS) image with a panchromatic (PAN) image. The main concern is how to effectively combine the rich spectral information of LRMS image with the abundant spatial information of PAN image. Recently, many methods based on deep learning have been proposed for the pansharpening task. However, these methods usually has two main drawbacks: 1) requiring HRMS for supervised learning; and 2) simply ignoring the latent relation between the MS and PAN image and fusing them directly. To solve these problems, we propose a novel unsupervised network based on learnable degradation processes, dubbed as LDP-Net. A reblurring block and a graying block are designed to learn the corresponding degradation processes, respectively. In addition, a novel hybrid loss function is proposed to constrain both spatial and spectral consistency between the pansharpened image and the PAN and LRMS images at different resolutions. Experiments on Worldview2 and Worldview3 images demonstrate that our proposed LDP-Net can fuse PAN and LRMS images effectively without the help of HRMS samples, achieving promising performance in terms of both qualitative visual effects and quantitative metrics.
Unsupervised PET Reconstruction from a Bayesian Perspective
Oct 29, 2021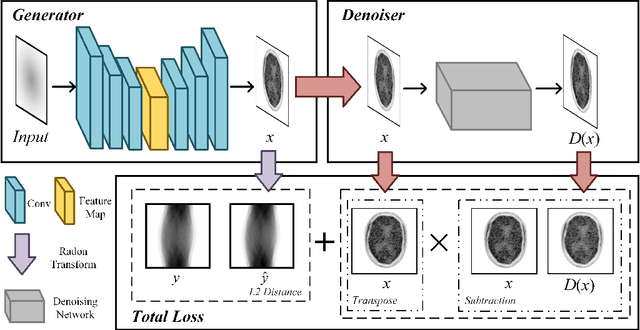

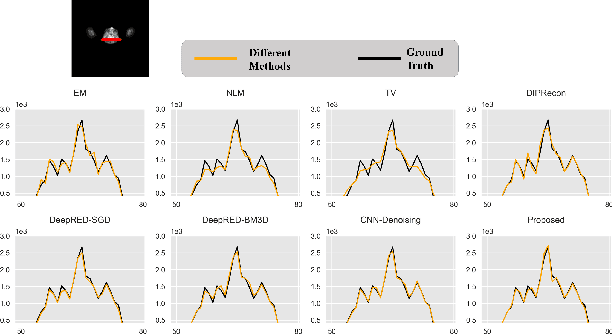
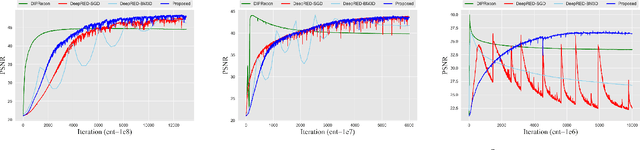
Abstract:Positron emission tomography (PET) reconstruction has become an ill-posed inverse problem due to low-count projection data, and a robust algorithm is urgently required to improve imaging quality. Recently, the deep image prior (DIP) has drawn much attention and has been successfully applied in several image restoration tasks, such as denoising and inpainting, since it does not need any labels (reference image). However, overfitting is a vital defect of this framework. Hence, many methods have been proposed to mitigate this problem, and DeepRED is a typical representation that combines DIP and regularization by denoising (RED). In this article, we leverage DeepRED from a Bayesian perspective to reconstruct PET images from a single corrupted sinogram without any supervised or auxiliary information. In contrast to the conventional denoisers customarily used in RED, a DnCNN-like denoiser, which can add an adaptive constraint to DIP and facilitate the computation of derivation, is employed. Moreover, to further enhance the regularization, Gaussian noise is injected into the gradient updates, deriving a Markov chain Monte Carlo (MCMC) sampler. Experimental studies on brain and whole-body datasets demonstrate that our proposed method can achieve better performance in terms of qualitative and quantitative results compared to several classic and state-of-the-art methods.
One Network to Solve Them All: A Sequential Multi-Task Joint Learning Network Framework for MR Imaging Pipeline
May 14, 2021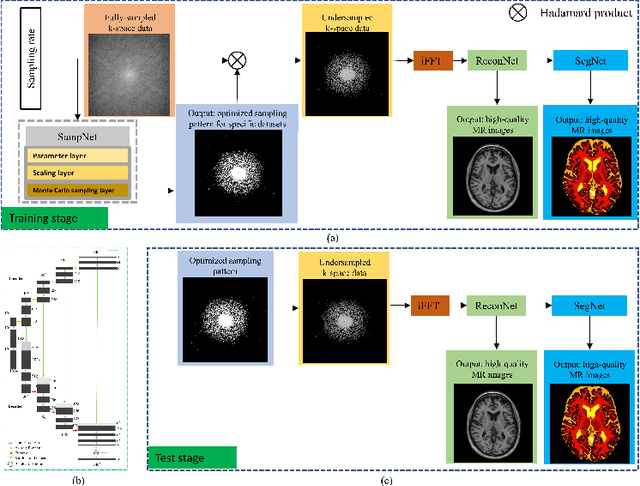
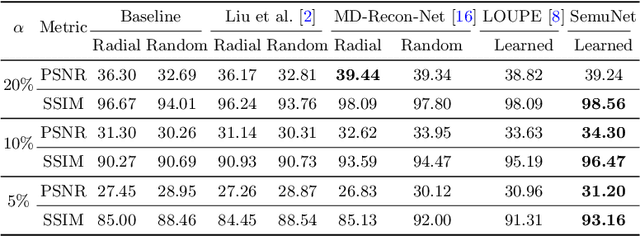
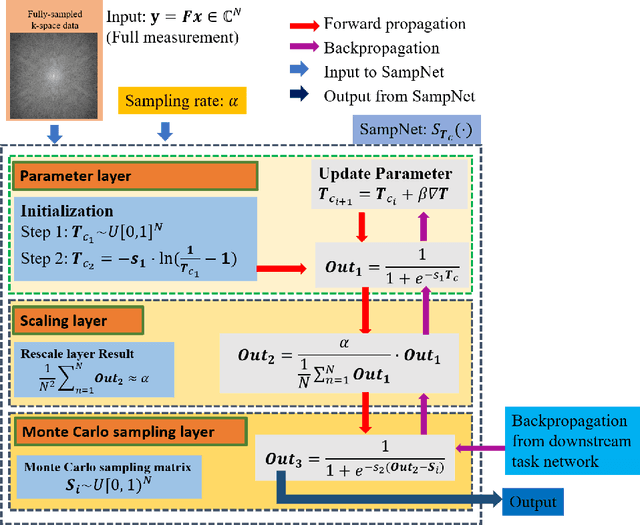
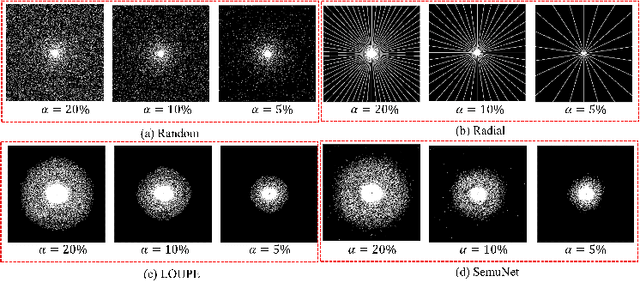
Abstract:Magnetic resonance imaging (MRI) acquisition, reconstruction, and segmentation are usually processed independently in the conventional practice of MRI workflow. It is easy to notice that there are significant relevances among these tasks and this procedure artificially cuts off these potential connections, which may lead to losing clinically important information for the final diagnosis. To involve these potential relations for further performance improvement, a sequential multi-task joint learning network model is proposed to train a combined end-to-end pipeline in a differentiable way, aiming at exploring the mutual influence among those tasks simultaneously. Our design consists of three cascaded modules: 1) deep sampling pattern learning module optimizes the $k$-space sampling pattern with predetermined sampling rate; 2) deep reconstruction module is dedicated to reconstructing MR images from the undersampled data using the learned sampling pattern; 3) deep segmentation module encodes MR images reconstructed from the previous module to segment the interested tissues. The proposed model retrieves the latently interactive and cyclic relations among those tasks, from which each task will be mutually beneficial. The proposed framework is verified on MRB dataset, which achieves superior performance on other SOTA methods in terms of both reconstruction and segmentation.
Deep Learning based Multi-modal Computing with Feature Disentanglement for MRI Image Synthesis
May 06, 2021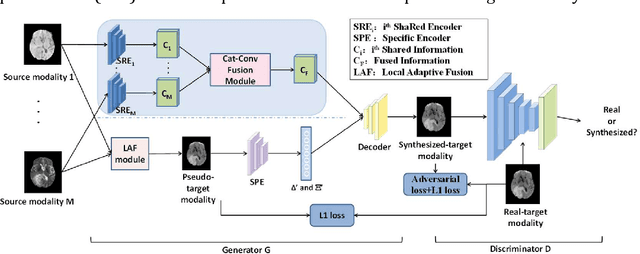
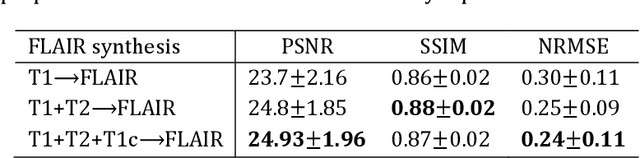
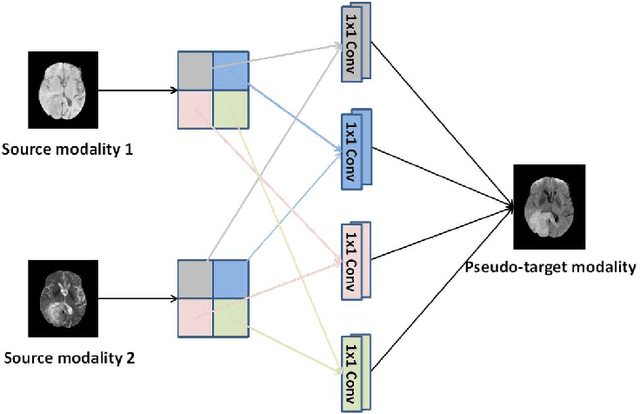
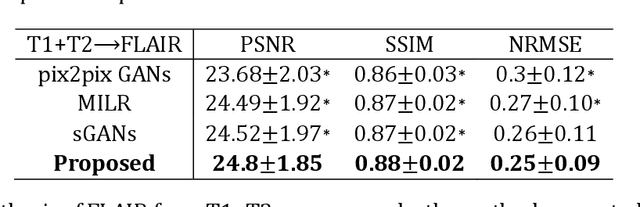
Abstract:Purpose: Different Magnetic resonance imaging (MRI) modalities of the same anatomical structure are required to present different pathological information from the physical level for diagnostic needs. However, it is often difficult to obtain full-sequence MRI images of patients owing to limitations such as time consumption and high cost. The purpose of this work is to develop an algorithm for target MRI sequences prediction with high accuracy, and provide more information for clinical diagnosis. Methods: We propose a deep learning based multi-modal computing model for MRI synthesis with feature disentanglement strategy. To take full advantage of the complementary information provided by different modalities, multi-modal MRI sequences are utilized as input. Notably, the proposed approach decomposes each input modality into modality-invariant space with shared information and modality-specific space with specific information, so that features are extracted separately to effectively process the input data. Subsequently, both of them are fused through the adaptive instance normalization (AdaIN) layer in the decoder. In addition, to address the lack of specific information of the target modality in the test phase, a local adaptive fusion (LAF) module is adopted to generate a modality-like pseudo-target with specific information similar to the ground truth. Results: To evaluate the synthesis performance, we verify our method on the BRATS2015 dataset of 164 subjects. The experimental results demonstrate our approach significantly outperforms the benchmark method and other state-of-the-art medical image synthesis methods in both quantitative and qualitative measures. Compared with the pix2pixGANs method, the PSNR improves from 23.68 to 24.8. Conclusion: The proposed method could be effective in prediction of target MRI sequences, and useful for clinical diagnosis and treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge