Cong Sun
Extracting Post-Acute Sequelae of SARS-CoV-2 Infection Symptoms from Clinical Notes via Hybrid Natural Language Processing
Aug 17, 2025Abstract:Accurately and efficiently diagnosing Post-Acute Sequelae of COVID-19 (PASC) remains challenging due to its myriad symptoms that evolve over long- and variable-time intervals. To address this issue, we developed a hybrid natural language processing pipeline that integrates rule-based named entity recognition with BERT-based assertion detection modules for PASC-symptom extraction and assertion detection from clinical notes. We developed a comprehensive PASC lexicon with clinical specialists. From 11 health systems of the RECOVER initiative network across the U.S., we curated 160 intake progress notes for model development and evaluation, and collected 47,654 progress notes for a population-level prevalence study. We achieved an average F1 score of 0.82 in one-site internal validation and 0.76 in 10-site external validation for assertion detection. Our pipeline processed each note at $2.448\pm 0.812$ seconds on average. Spearman correlation tests showed $\rho >0.83$ for positive mentions and $\rho >0.72$ for negative ones, both with $P <0.0001$. These demonstrate the effectiveness and efficiency of our models and their potential for improving PASC diagnosis.
Generative Large Language Models Trained for Detecting Errors in Radiology Reports
Apr 06, 2025Abstract:In this retrospective study, a dataset was constructed with two parts. The first part included 1,656 synthetic chest radiology reports generated by GPT-4 using specified prompts, with 828 being error-free synthetic reports and 828 containing errors. The second part included 614 reports: 307 error-free reports between 2011 and 2016 from the MIMIC-CXR database and 307 corresponding synthetic reports with errors generated by GPT-4 on the basis of these MIMIC-CXR reports and specified prompts. All errors were categorized into four types: negation, left/right, interval change, and transcription errors. Then, several models, including Llama-3, GPT-4, and BiomedBERT, were refined using zero-shot prompting, few-shot prompting, or fine-tuning strategies. Finally, the performance of these models was evaluated using the F1 score, 95\% confidence interval (CI) and paired-sample t-tests on our constructed dataset, with the prediction results further assessed by radiologists. Using zero-shot prompting, the fine-tuned Llama-3-70B-Instruct model achieved the best performance with the following F1 scores: 0.769 for negation errors, 0.772 for left/right errors, 0.750 for interval change errors, 0.828 for transcription errors, and 0.780 overall. In the real-world evaluation phase, two radiologists reviewed 200 randomly selected reports output by the model. Of these, 99 were confirmed to contain errors detected by the models by both radiologists, and 163 were confirmed to contain model-detected errors by at least one radiologist. Generative LLMs, fine-tuned on synthetic and MIMIC-CXR radiology reports, greatly enhanced error detection in radiology reports.
AtlasSeg: Atlas Prior Guided Dual-U-Net for Cortical Segmentation in Fetal Brain MRI
Nov 05, 2024



Abstract:Accurate tissue segmentation in fetal brain MRI remains challenging due to the dynamically changing anatomical anatomy and contrast during fetal development. To enhance segmentation accuracy throughout gestation, we introduced AtlasSeg, a dual-U-shape convolution network incorporating gestational age (GA) specific information as guidance. By providing a publicly available fetal brain atlas with segmentation label at the corresponding GA, AtlasSeg effectively extracted the contextual features of age-specific patterns in atlas branch and generated tissue segmentation in segmentation branch. Multi-scale attentive atlas feature fusions were constructed in all stages during encoding and decoding, giving rise to a dual-U-shape network to assist feature flow and information interactions between two branches. AtlasSeg outperformed six well-known segmentation networks in both our internal fetal brain MRI dataset and the external FeTA dataset. Ablation experiments demonstrate the efficiency of atlas guidance and the attention mechanism. The proposed AtlasSeg demonstrated superior segmentation performance against other convolution networks with higher segmentation accuracy, and may facilitate fetal brain MRI analysis in large-scale fetal brain studies.
A Literature Review and Framework for Human Evaluation of Generative Large Language Models in Healthcare
May 04, 2024Abstract:As generative artificial intelligence (AI), particularly Large Language Models (LLMs), continues to permeate healthcare, it remains crucial to supplement traditional automated evaluations with human expert evaluation. Understanding and evaluating the generated texts is vital for ensuring safety, reliability, and effectiveness. However, the cumbersome, time-consuming, and non-standardized nature of human evaluation presents significant obstacles to the widespread adoption of LLMs in practice. This study reviews existing literature on human evaluation methodologies for LLMs within healthcare. We highlight a notable need for a standardized and consistent human evaluation approach. Our extensive literature search, adhering to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, spans publications from January 2018 to February 2024. This review provides a comprehensive overview of the human evaluation approaches used in diverse healthcare applications.This analysis examines the human evaluation of LLMs across various medical specialties, addressing factors such as evaluation dimensions, sample types, and sizes, the selection and recruitment of evaluators, frameworks and metrics, the evaluation process, and statistical analysis of the results. Drawing from diverse evaluation strategies highlighted in these studies, we propose a comprehensive and practical framework for human evaluation of generative LLMs, named QUEST: Quality of Information, Understanding and Reasoning, Expression Style and Persona, Safety and Harm, and Trust and Confidence. This framework aims to improve the reliability, generalizability, and applicability of human evaluation of generative LLMs in different healthcare applications by defining clear evaluation dimensions and offering detailed guidelines.
Deep learning with noisy labels in medical prediction problems: a scoping review
Mar 19, 2024



Abstract:Objectives: Medical research faces substantial challenges from noisy labels attributed to factors like inter-expert variability and machine-extracted labels. Despite this, the adoption of label noise management remains limited, and label noise is largely ignored. To this end, there is a critical need to conduct a scoping review focusing on the problem space. This scoping review aims to comprehensively review label noise management in deep learning-based medical prediction problems, which includes label noise detection, label noise handling, and evaluation. Research involving label uncertainty is also included. Methods: Our scoping review follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. We searched 4 databases, including PubMed, IEEE Xplore, Google Scholar, and Semantic Scholar. Our search terms include "noisy label AND medical / healthcare / clinical", "un-certainty AND medical / healthcare / clinical", and "noise AND medical / healthcare / clinical". Results: A total of 60 papers met inclusion criteria between 2016 and 2023. A series of practical questions in medical research are investigated. These include the sources of label noise, the impact of label noise, the detection of label noise, label noise handling techniques, and their evaluation. Categorization of both label noise detection methods and handling techniques are provided. Discussion: From a methodological perspective, we observe that the medical community has been up to date with the broader deep-learning community, given that most techniques have been evaluated on medical data. We recommend considering label noise as a standard element in medical research, even if it is not dedicated to handling noisy labels. Initial experiments can start with easy-to-implement methods, such as noise-robust loss functions, weighting, and curriculum learning.
CafkNet: GNN-Empowered Forward Kinematic Modeling for Cable-Driven Parallel Robots
Mar 05, 2024



Abstract:The Cable-Driven Parallel Robots (CDPRs) have gained significant attention due to their high payload capacity and large workspace. When deploying CDPRs in practice, one of the challenges is kinematic modeling. Unlike serial mechanisms, CDPRs have a simple inverse kinematics problem but a complex forward kinematics (FK) issue. Therefore, the development of accurate and efficient FK solvers has been a prominent research focus in CDPR applications. By observing the topology within CDPRs, in this paper, we propose a graph-based representation to model CDPRs and introduce CafkNet, a fast and general FK solver, leveraging Graph Neural Network (GNN). CafkNet is extensively tested on 3D and 2D CDPRs in different configurations, both in simulators and real scenarios. The results demonstrate its ability to learn CDPRs' internal topology and accurately solve the FK problem. Then, the zero-shot generalization from one configuration to another is validated. Also, the sim2real gap can be bridged by CafkNet using both simulation and real-world data. To the best of our knowledge, it is the first study that employs the GNN to solve FK problem for CDPRs.
Demonstration-based learning for few-shot biomedical named entity recognition under machine reading comprehension
Aug 12, 2023



Abstract:Although deep learning techniques have shown significant achievements, they frequently depend on extensive amounts of hand-labeled data and tend to perform inadequately in few-shot scenarios. The objective of this study is to devise a strategy that can improve the model's capability to recognize biomedical entities in scenarios of few-shot learning. By redefining biomedical named entity recognition (BioNER) as a machine reading comprehension (MRC) problem, we propose a demonstration-based learning method to address few-shot BioNER, which involves constructing appropriate task demonstrations. In assessing our proposed method, we compared the proposed method with existing advanced methods using six benchmark datasets, including BC4CHEMD, BC5CDR-Chemical, BC5CDR-Disease, NCBI-Disease, BC2GM, and JNLPBA. We examined the models' efficacy by reporting F1 scores from both the 25-shot and 50-shot learning experiments. In 25-shot learning, we observed 1.1% improvements in the average F1 scores compared to the baseline method, reaching 61.7%, 84.1%, 69.1%, 70.1%, 50.6%, and 59.9% on six datasets, respectively. In 50-shot learning, we further improved the average F1 scores by 1.0% compared to the baseline method, reaching 73.1%, 86.8%, 76.1%, 75.6%, 61.7%, and 65.4%, respectively. We reported that in the realm of few-shot learning BioNER, MRC-based language models are much more proficient in recognizing biomedical entities compared to the sequence labeling approach. Furthermore, our MRC-language models can compete successfully with fully-supervised learning methodologies that rely heavily on the availability of abundant annotated data. These results highlight possible pathways for future advancements in few-shot BioNER methodologies.
A Motion Assessment Method for Reference Stack Selection in Fetal Brain MRI Reconstruction Based on Tensor Rank Approximation
Jun 30, 2023



Abstract:Purpose: Slice-to-volume registration and super-resolution reconstruction (SVR-SRR) is commonly used to generate 3D volumes of the fetal brain from 2D stacks of slices acquired in multiple orientations. A critical initial step in this pipeline is to select one stack with the minimum motion as a reference for registration. An accurate and unbiased motion assessment (MA) is thus crucial for successful selection. Methods: We presented a MA method that determines the minimum motion stack based on 3D low-rank approximation using CANDECOMP/PARAFAC (CP) decomposition. Compared to the current 2D singular value decomposition (SVD) based method that requires flattening stacks into matrices to obtain ranks, in which the spatial information is lost, the CP-based method can factorize 3D stack into low-rank and sparse components in a computationally efficient manner. The difference between the original stack and its low-rank approximation was proposed as the motion indicator. Results: Compared to SVD-based methods, our proposed CP-based MA demonstrated higher sensitivity in detecting small motion with a lower baseline bias. Experiments on randomly simulated motion illustrated that the proposed CP method achieved a higher success rate of 95.45% in identifying the minimum motion stack, compared to SVD-based method with a success rate of 58.18%. We further demonstrated that combining CP-based MA with existing SRR-SVR pipeline significantly improved 3D volume reconstruction. Conclusion: The proposed CP-based MA method showed superior performance compared to SVD-based methods with higher sensitivity to motion, success rate, and lower baseline bias, and can be used as a prior step to improve fetal brain reconstruction.
A microstructure estimation Transformer inspired by sparse representation for diffusion MRI
May 13, 2022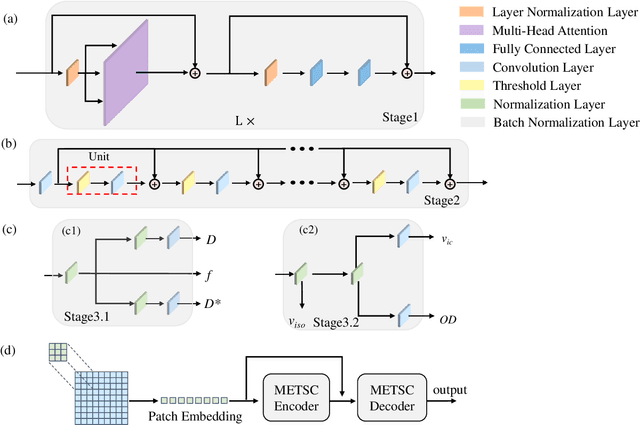
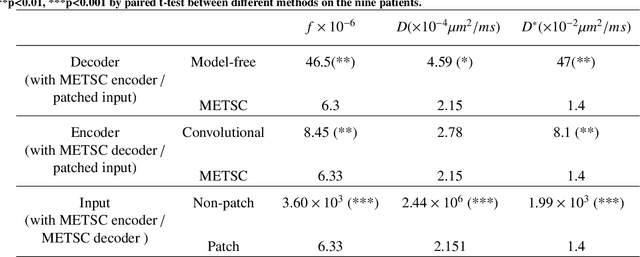
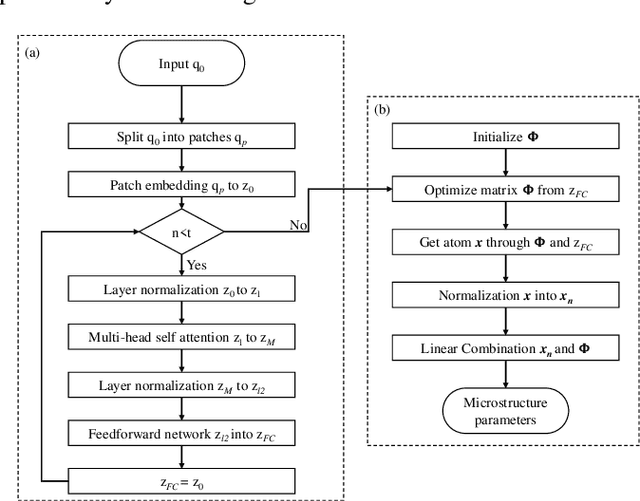
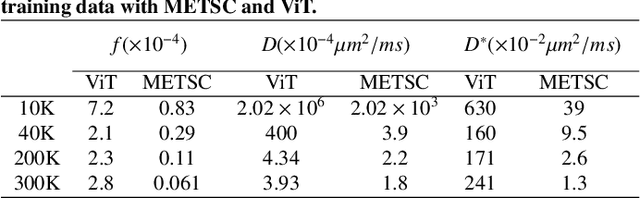
Abstract:Diffusion magnetic resonance imaging (dMRI) is an important tool in characterizing tissue microstructure based on biophysical models, which are complex and highly non-linear. Resolving microstructures with optimization techniques is prone to estimation errors and requires dense sampling in the q-space. Deep learning based approaches have been proposed to overcome these limitations. Motivated by the superior performance of the Transformer, in this work, we present a learning-based framework based on Transformer, namely, a Microstructure Estimation Transformer with Sparse Coding (METSC) for dMRI-based microstructure estimation with downsampled q-space data. To take advantage of the Transformer while addressing its limitation in large training data requirements, we explicitly introduce an inductive bias - model bias into the Transformer using a sparse coding technique to facilitate the training process. Thus, the METSC is composed with three stages, an embedding stage, a sparse representation stage, and a mapping stage. The embedding stage is a Transformer-based structure that encodes the signal to ensure the voxel is represented effectively. In the sparse representation stage, a dictionary is constructed by solving a sparse reconstruction problem that unfolds the Iterative Hard Thresholding (IHT) process. The mapping stage is essentially a decoder that computes the microstructural parameters from the output of the second stage, based on the weighted sum of normalized dictionary coefficients where the weights are also learned. We tested our framework on two dMRI models with downsampled q-space data, including the intravoxel incoherent motion (IVIM) model and the neurite orientation dispersion and density imaging (NODDI) model. The proposed method achieved up to 11.25 folds of acceleration in scan time and outperformed the other state-of-the-art learning-based methods.
AFFIRM: Affinity Fusion-based Framework for Iteratively Random Motion correction of multi-slice fetal brain MRI
May 12, 2022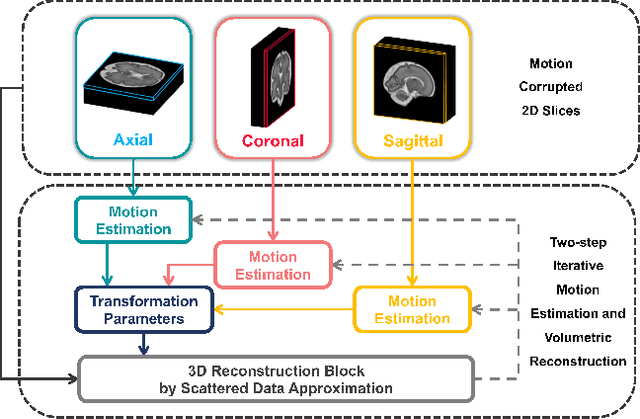
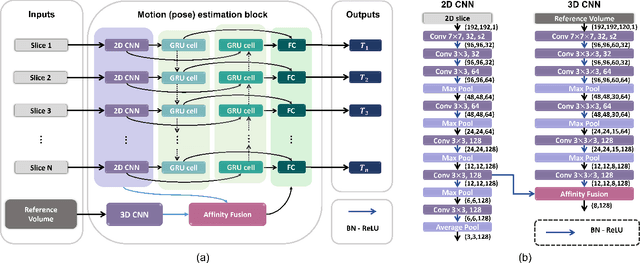
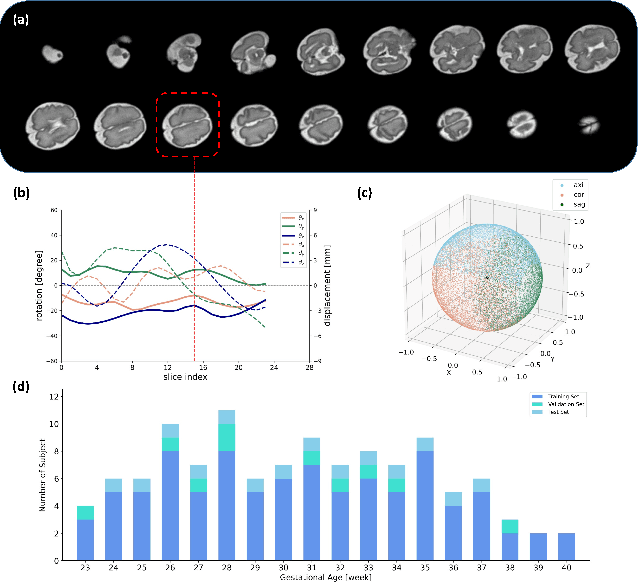
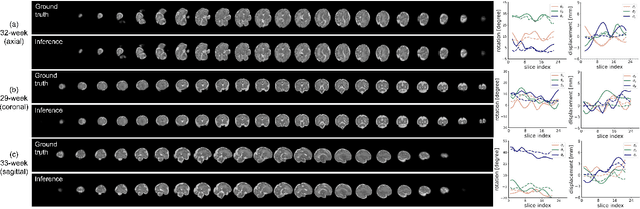
Abstract:Multi-slice magnetic resonance images of the fetal brain are usually contaminated by severe and arbitrary fetal and maternal motion. Hence, stable and robust motion correction is necessary to reconstruct high-resolution 3D fetal brain volume for clinical diagnosis and quantitative analysis. However, the conventional registration-based correction has a limited capture range and is insufficient for detecting relatively large motions. Here, we present a novel Affinity Fusion-based Framework for Iteratively Random Motion (AFFIRM) correction of the multi-slice fetal brain MRI. It learns the sequential motion from multiple stacks of slices and integrates the features between 2D slices and reconstructed 3D volume using affinity fusion, which resembles the iterations between slice-to-volume registration and volumetric reconstruction in the regular pipeline. The method accurately estimates the motion regardless of brain orientations and outperforms other state-of-the-art learning-based methods on the simulated motion-corrupted data, with a 48.4% reduction of mean absolute error for rotation and 61.3% for displacement. We then incorporated AFFIRM into the multi-resolution slice-to-volume registration and tested it on the real-world fetal MRI scans at different gestation stages. The results indicated that adding AFFIRM to the conventional pipeline improved the success rate of fetal brain super-resolution reconstruction from 77.2% to 91.9%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge