Chu Han
A Standardized Pipeline for Colon Nuclei Identification and Counting Challenge
Mar 20, 2022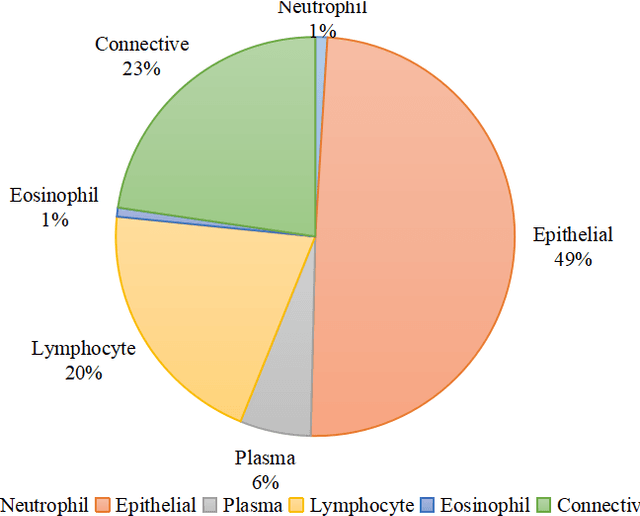
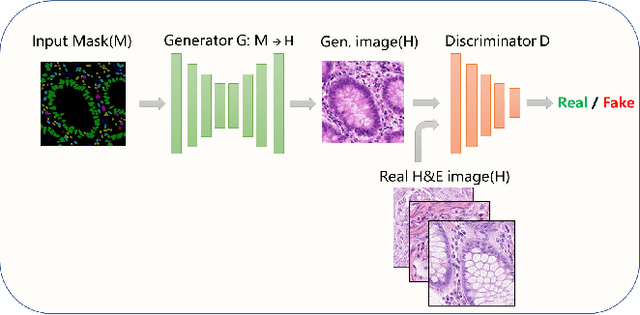
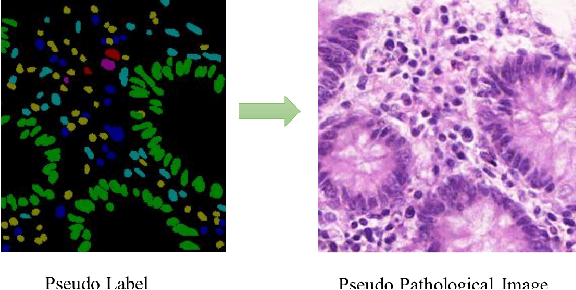
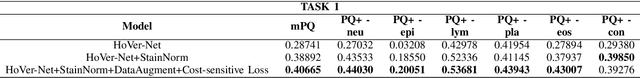
Abstract:Nuclear segmentation and classification is an essential step for computational pathology. TIA lab from Warwick University organized a nuclear segmentation and classification challenge (CoNIC) for H&E stained histopathology images in colorectal cancer with two highly correlated tasks, nuclei segmentation and classification task and cellular composition task. There are a few obstacles we have to address in this challenge, 1) limited training samples, 2) color variation, 3) imbalanced annotations, 4) similar morphological appearance among classes. To deal with these challenges, we proposed a standardized pipeline for nuclear segmentation and classification by integrating several pluggable components. First, we built a GAN-based model to automatically generate pseudo images for data augmentation. Then we trained a self-supervised stain normalization model to solve the color variation problem. Next we constructed a baseline model HoVer-Net with cost-sensitive loss to encourage the model pay more attention on the minority classes. According to the results of the leaderboard, our proposed pipeline achieves 0.40665 mPQ+ (Rank 49th) and 0.62199 r2 (Rank 10th) in the preliminary test phase.
RestainNet: a self-supervised digital re-stainer for stain normalization
Feb 28, 2022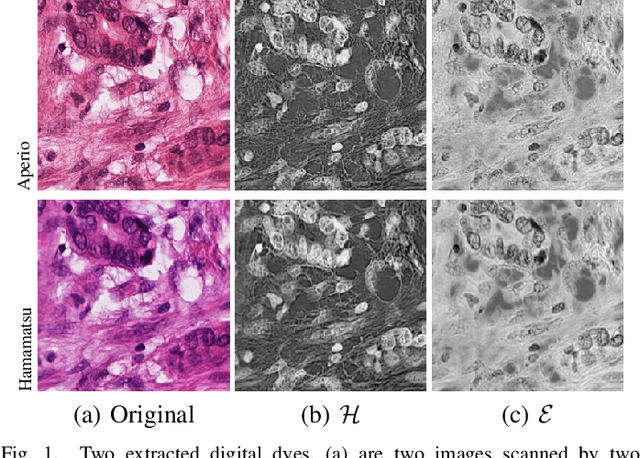
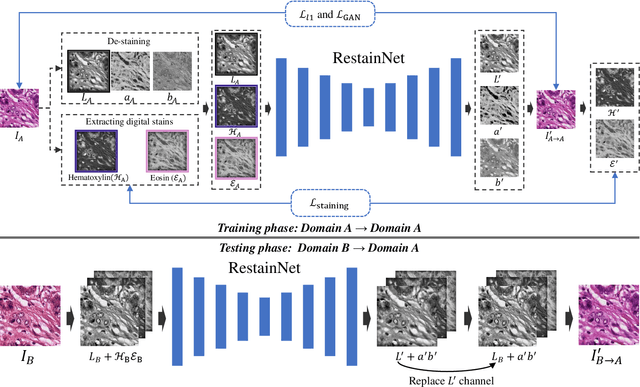
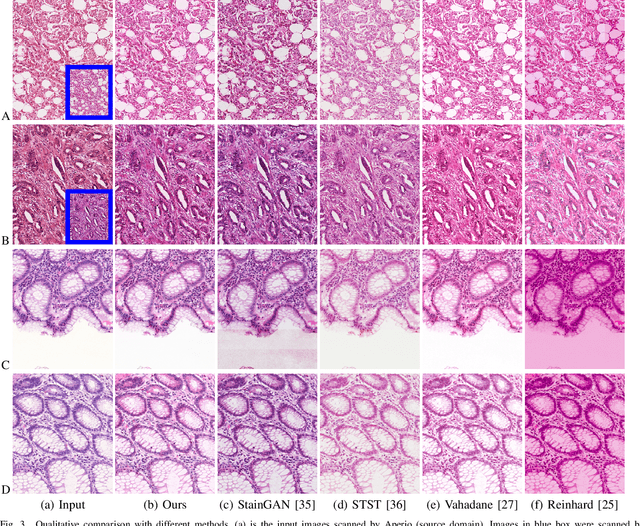
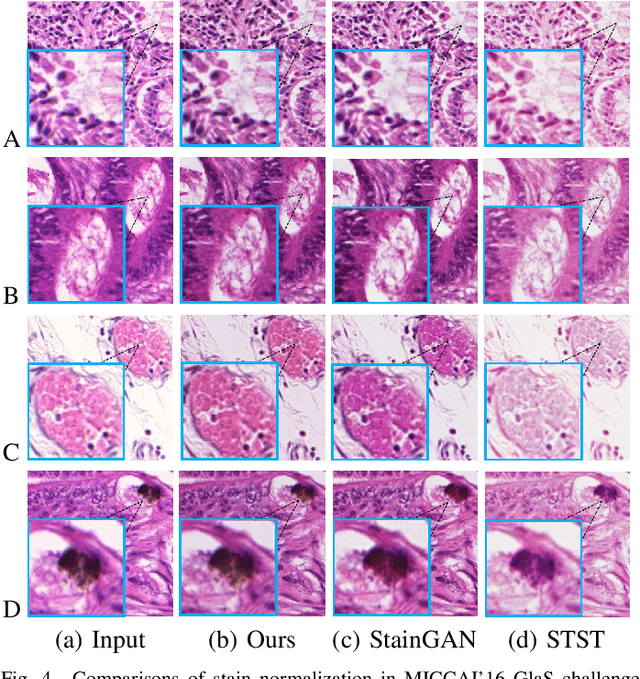
Abstract:Color inconsistency is an inevitable challenge in computational pathology, which generally happens because of stain intensity variations or sections scanned by different scanners. It harms the pathological image analysis methods, especially the learning-based models. A series of approaches have been proposed for stain normalization. However, most of them are lack flexibility in practice. In this paper, we formulated stain normalization as a digital re-staining process and proposed a self-supervised learning model, which is called RestainNet. Our network is regarded as a digital restainer which learns how to re-stain an unstained (grayscale) image. Two digital stains, Hematoxylin (H) and Eosin (E) were extracted from the original image by Beer-Lambert's Law. We proposed a staining loss to maintain the correctness of stain intensity during the restaining process. Thanks to the self-supervised nature, paired training samples are no longer necessary, which demonstrates great flexibility in practical usage. Our RestainNet outperforms existing approaches and achieves state-of-the-art performance with regard to color correctness and structure preservation. We further conducted experiments on the segmentation and classification tasks and the proposed RestainNet achieved outstanding performance compared with SOTA methods. The self-supervised design allows the network to learn any staining style with no extra effort.
PDBL: Improving Histopathological Tissue Classification with Plug-and-Play Pyramidal Deep-Broad Learning
Nov 04, 2021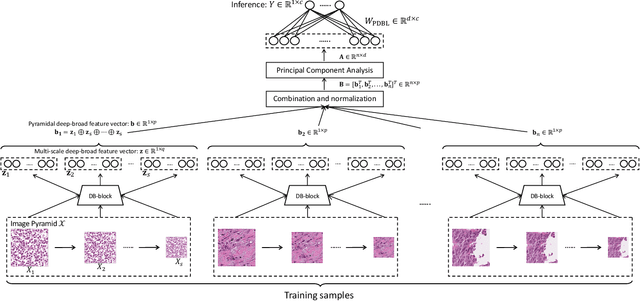
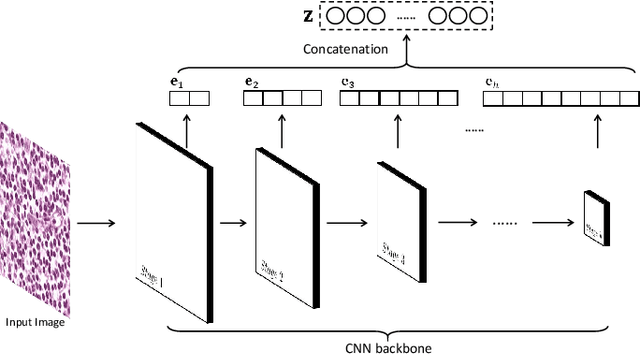

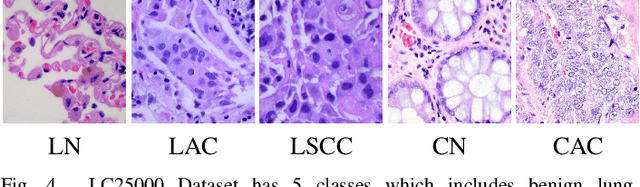
Abstract:Histopathological tissue classification is a fundamental task in pathomics cancer research. Precisely differentiating different tissue types is a benefit for the downstream researches, like cancer diagnosis, prognosis and etc. Existing works mostly leverage the popular classification backbones in computer vision to achieve histopathological tissue classification. In this paper, we proposed a super lightweight plug-and-play module, named Pyramidal Deep-Broad Learning (PDBL), for any well-trained classification backbone to further improve the classification performance without a re-training burden. We mimic how pathologists observe pathology slides in different magnifications and construct an image pyramid for the input image in order to obtain the pyramidal contextual information. For each level in the pyramid, we extract the multi-scale deep-broad features by our proposed Deep-Broad block (DB-block). We equipped PDBL in three popular classification backbones, ShuffLeNetV2, EfficientNetb0, and ResNet50 to evaluate the effectiveness and efficiency of our proposed module on two datasets (Kather Multiclass Dataset and the LC25000 Dataset). Experimental results demonstrate the proposed PDBL can steadily improve the tissue-level classification performance for any CNN backbones, especially for the lightweight models when given a small among of training samples (less than 10%), which greatly saves the computational time and annotation efforts.
Multi-Layer Pseudo-Supervision for Histopathology Tissue Semantic Segmentation using Patch-level Classification Labels
Oct 14, 2021

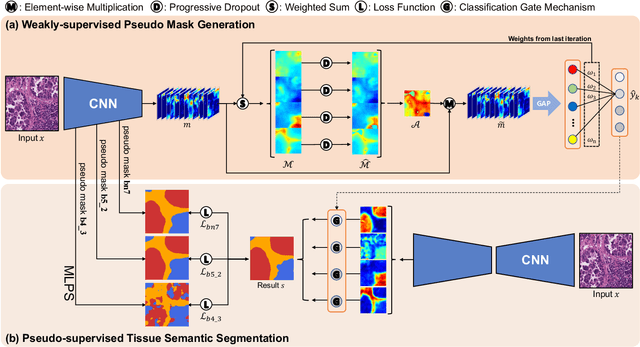
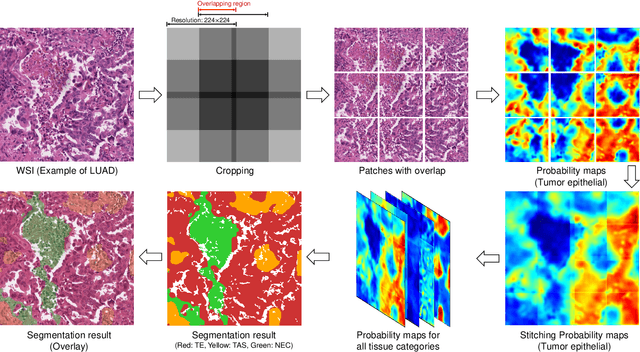
Abstract:Tissue-level semantic segmentation is a vital step in computational pathology. Fully-supervised models have already achieved outstanding performance with dense pixel-level annotations. However, drawing such labels on the giga-pixel whole slide images is extremely expensive and time-consuming. In this paper, we use only patch-level classification labels to achieve tissue semantic segmentation on histopathology images, finally reducing the annotation efforts. We proposed a two-step model including a classification and a segmentation phases. In the classification phase, we proposed a CAM-based model to generate pseudo masks by patch-level labels. In the segmentation phase, we achieved tissue semantic segmentation by our proposed Multi-Layer Pseudo-Supervision. Several technical novelties have been proposed to reduce the information gap between pixel-level and patch-level annotations. As a part of this paper, we introduced a new weakly-supervised semantic segmentation (WSSS) dataset for lung adenocarcinoma (LUAD-HistoSeg). We conducted several experiments to evaluate our proposed model on two datasets. Our proposed model outperforms two state-of-the-art WSSS approaches. Note that we can achieve comparable quantitative and qualitative results with the fully-supervised model, with only around a 2\% gap for MIoU and FwIoU. By comparing with manual labeling, our model can greatly save the annotation time from hours to minutes. The source code is available at: \url{https://github.com/ChuHan89/WSSS-Tissue}.
TENet: Triple Excitation Network for Video Salient Object Detection
Aug 30, 2020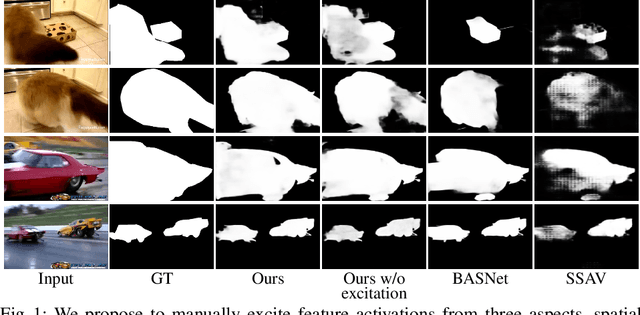
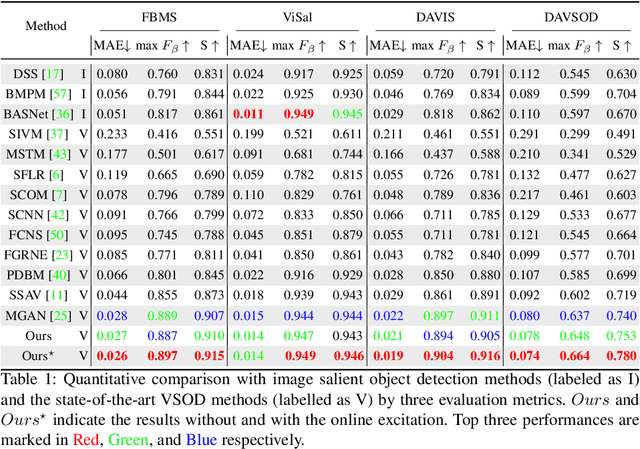
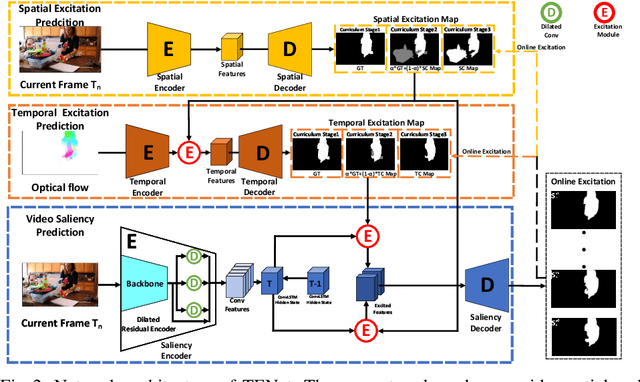
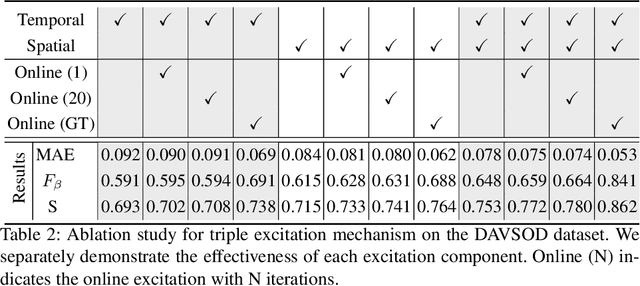
Abstract:In this paper, we propose a simple yet effective approach, named Triple Excitation Network, to reinforce the training of video salient object detection (VSOD) from three aspects, spatial, temporal, and online excitations. These excitation mechanisms are designed following the spirit of curriculum learning and aim to reduce learning ambiguities at the beginning of training by selectively exciting feature activations using ground truth. Then we gradually reduce the weight of ground truth excitations by a curriculum rate and replace it by a curriculum complementary map for better and faster convergence. In particular, the spatial excitation strengthens feature activations for clear object boundaries, while the temporal excitation imposes motions to emphasize spatio-temporal salient regions. Spatial and temporal excitations can combat the saliency shifting problem and conflict between spatial and temporal features of VSOD. Furthermore, our semi-curriculum learning design enables the first online refinement strategy for VSOD, which allows exciting and boosting saliency responses during testing without re-training. The proposed triple excitations can easily plug in different VSOD methods. Extensive experiments show the effectiveness of all three excitation methods and the proposed method outperforms state-of-the-art image and video salient object detection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge