Jijun Cheng
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
A Standardized Pipeline for Colon Nuclei Identification and Counting Challenge
Mar 20, 2022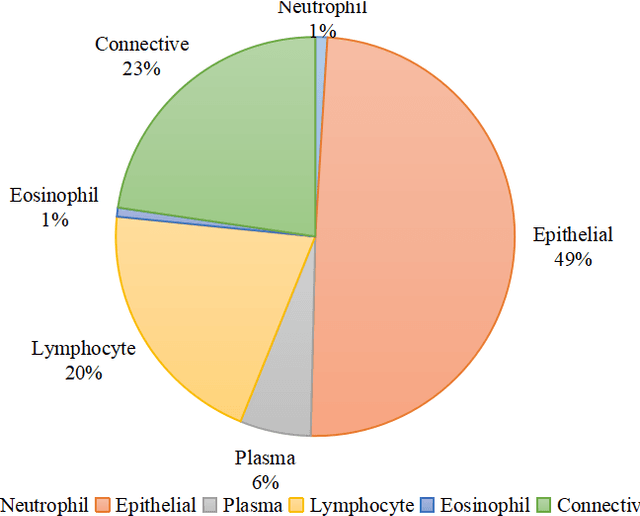
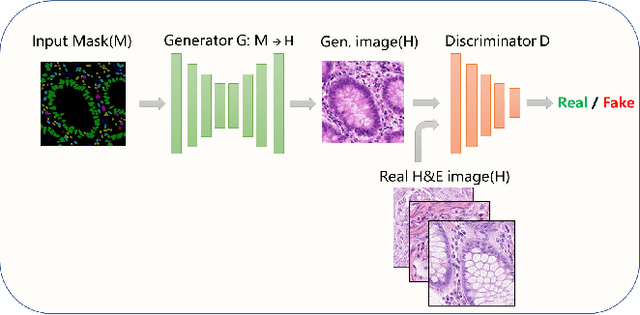
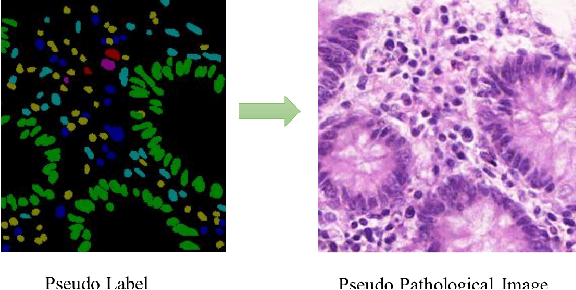
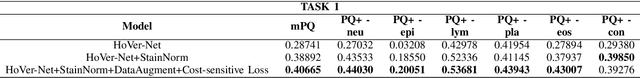
Abstract:Nuclear segmentation and classification is an essential step for computational pathology. TIA lab from Warwick University organized a nuclear segmentation and classification challenge (CoNIC) for H&E stained histopathology images in colorectal cancer with two highly correlated tasks, nuclei segmentation and classification task and cellular composition task. There are a few obstacles we have to address in this challenge, 1) limited training samples, 2) color variation, 3) imbalanced annotations, 4) similar morphological appearance among classes. To deal with these challenges, we proposed a standardized pipeline for nuclear segmentation and classification by integrating several pluggable components. First, we built a GAN-based model to automatically generate pseudo images for data augmentation. Then we trained a self-supervised stain normalization model to solve the color variation problem. Next we constructed a baseline model HoVer-Net with cost-sensitive loss to encourage the model pay more attention on the minority classes. According to the results of the leaderboard, our proposed pipeline achieves 0.40665 mPQ+ (Rank 49th) and 0.62199 r2 (Rank 10th) in the preliminary test phase.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge