Chenhui Wang
UCLA Department of Statistics, Los Angeles, CA
Dynamic Differential Linear Attention: Enhancing Linear Diffusion Transformer for High-Quality Image Generation
Jan 20, 2026Abstract:Diffusion transformers (DiTs) have emerged as a powerful architecture for high-fidelity image generation, yet the quadratic cost of self-attention poses a major scalability bottleneck. To address this, linear attention mechanisms have been adopted to reduce computational cost; unfortunately, the resulting linear diffusion transformers (LiTs) models often come at the expense of generative performance, frequently producing over-smoothed attention weights that limit expressiveness. In this work, we introduce Dynamic Differential Linear Attention (DyDiLA), a novel linear attention formulation that enhances the effectiveness of LiTs by mitigating the oversmoothing issue and improving generation quality. Specifically, the novelty of DyDiLA lies in three key designs: (i) dynamic projection module, which facilitates the decoupling of token representations by learning with dynamically assigned knowledge; (ii) dynamic measure kernel, which provides a better similarity measurement to capture fine-grained semantic distinctions between tokens by dynamically assigning kernel functions for token processing; and (iii) token differential operator, which enables more robust query-to-key retrieval by calculating the differences between the tokens and their corresponding information redundancy produced by dynamic measure kernel. To capitalize on DyDiLA, we introduce a refined LiT, termed DyDi-LiT, that systematically incorporates our advancements. Extensive experiments show that DyDi-LiT consistently outperforms current state-of-the-art (SOTA) models across multiple metrics, underscoring its strong practical potential.
OneRec-V2 Technical Report
Aug 28, 2025



Abstract:Recent breakthroughs in generative AI have transformed recommender systems through end-to-end generation. OneRec reformulates recommendation as an autoregressive generation task, achieving high Model FLOPs Utilization. While OneRec-V1 has shown significant empirical success in real-world deployment, two critical challenges hinder its scalability and performance: (1) inefficient computational allocation where 97.66% of resources are consumed by sequence encoding rather than generation, and (2) limitations in reinforcement learning relying solely on reward models. To address these challenges, we propose OneRec-V2, featuring: (1) Lazy Decoder-Only Architecture: Eliminates encoder bottlenecks, reducing total computation by 94% and training resources by 90%, enabling successful scaling to 8B parameters. (2) Preference Alignment with Real-World User Interactions: Incorporates Duration-Aware Reward Shaping and Adaptive Ratio Clipping to better align with user preferences using real-world feedback. Extensive A/B tests on Kuaishou demonstrate OneRec-V2's effectiveness, improving App Stay Time by 0.467%/0.741% while balancing multi-objective recommendations. This work advances generative recommendation scalability and alignment with real-world feedback, representing a step forward in the development of end-to-end recommender systems.
LangMamba: A Language-driven Mamba Framework for Low-dose CT Denoising with Vision-language Models
Jul 08, 2025Abstract:Low-dose computed tomography (LDCT) reduces radiation exposure but often degrades image quality, potentially compromising diagnostic accuracy. Existing deep learning-based denoising methods focus primarily on pixel-level mappings, overlooking the potential benefits of high-level semantic guidance. Recent advances in vision-language models (VLMs) suggest that language can serve as a powerful tool for capturing structured semantic information, offering new opportunities to improve LDCT reconstruction. In this paper, we introduce LangMamba, a Language-driven Mamba framework for LDCT denoising that leverages VLM-derived representations to enhance supervision from normal-dose CT (NDCT). LangMamba follows a two-stage learning strategy. First, we pre-train a Language-guided AutoEncoder (LangAE) that leverages frozen VLMs to map NDCT images into a semantic space enriched with anatomical information. Second, we synergize LangAE with two key components to guide LDCT denoising: Semantic-Enhanced Efficient Denoiser (SEED), which enhances NDCT-relevant local semantic while capturing global features with efficient Mamba mechanism, and Language-engaged Dual-space Alignment (LangDA) Loss, which ensures that denoised images align with NDCT in both perceptual and semantic spaces. Extensive experiments on two public datasets demonstrate that LangMamba outperforms conventional state-of-the-art methods, significantly improving detail preservation and visual fidelity. Remarkably, LangAE exhibits strong generalizability to unseen datasets, thereby reducing training costs. Furthermore, LangDA loss improves explainability by integrating language-guided insights into image reconstruction and offers a plug-and-play fashion. Our findings shed new light on the potential of language as a supervisory signal to advance LDCT denoising. The code is publicly available on https://github.com/hao1635/LangMamba.
OneRec Technical Report
Jun 16, 2025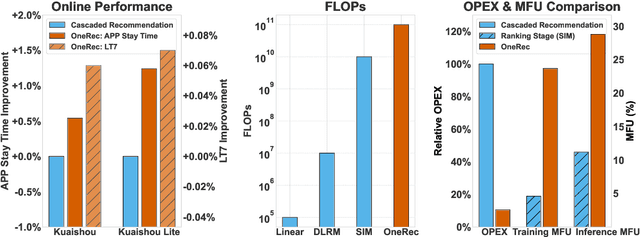

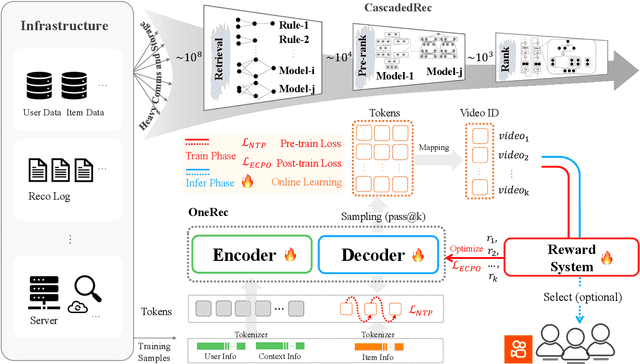
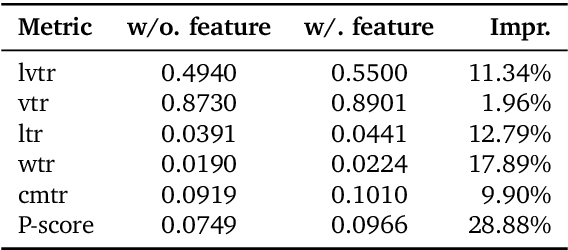
Abstract:Recommender systems have been widely used in various large-scale user-oriented platforms for many years. However, compared to the rapid developments in the AI community, recommendation systems have not achieved a breakthrough in recent years. For instance, they still rely on a multi-stage cascaded architecture rather than an end-to-end approach, leading to computational fragmentation and optimization inconsistencies, and hindering the effective application of key breakthrough technologies from the AI community in recommendation scenarios. To address these issues, we propose OneRec, which reshapes the recommendation system through an end-to-end generative approach and achieves promising results. Firstly, we have enhanced the computational FLOPs of the current recommendation model by 10 $\times$ and have identified the scaling laws for recommendations within certain boundaries. Secondly, reinforcement learning techniques, previously difficult to apply for optimizing recommendations, show significant potential in this framework. Lastly, through infrastructure optimizations, we have achieved 23.7% and 28.8% Model FLOPs Utilization (MFU) on flagship GPUs during training and inference, respectively, aligning closely with the LLM community. This architecture significantly reduces communication and storage overhead, resulting in operating expense that is only 10.6% of traditional recommendation pipelines. Deployed in Kuaishou/Kuaishou Lite APP, it handles 25% of total queries per second, enhancing overall App Stay Time by 0.54% and 1.24%, respectively. Additionally, we have observed significant increases in metrics such as 7-day Lifetime, which is a crucial indicator of recommendation experience. We also provide practical lessons and insights derived from developing, optimizing, and maintaining a production-scale recommendation system with significant real-world impact.
MedITok: A Unified Tokenizer for Medical Image Synthesis and Interpretation
May 25, 2025Abstract:Advanced autoregressive models have reshaped multimodal AI. However, their transformative potential in medical imaging remains largely untapped due to the absence of a unified visual tokenizer -- one capable of capturing fine-grained visual structures for faithful image reconstruction and realistic image synthesis, as well as rich semantics for accurate diagnosis and image interpretation. To this end, we present MedITok, the first unified tokenizer tailored for medical images, encoding both low-level structural details and high-level clinical semantics within a unified latent space. To balance these competing objectives, we introduce a novel two-stage training framework: a visual representation alignment stage that cold-starts the tokenizer reconstruction learning with a visual semantic constraint, followed by a textual semantic representation alignment stage that infuses detailed clinical semantics into the latent space. Trained on the meticulously collected large-scale dataset with over 30 million medical images and 2 million image-caption pairs, MedITok achieves state-of-the-art performance on more than 30 datasets across 9 imaging modalities and 4 different tasks. By providing a unified token space for autoregressive modeling, MedITok supports a wide range of tasks in clinical diagnostics and generative healthcare applications. Model and code will be made publicly available at: https://github.com/Masaaki-75/meditok.
Autoregressive Medical Image Segmentation via Next-Scale Mask Prediction
Feb 28, 2025



Abstract:While deep learning has significantly advanced medical image segmentation, most existing methods still struggle with handling complex anatomical regions. Cascaded or deep supervision-based approaches attempt to address this challenge through multi-scale feature learning but fail to establish sufficient inter-scale dependencies, as each scale relies solely on the features of the immediate predecessor. To this end, we propose the AutoRegressive Segmentation framework via next-scale mask prediction, termed AR-Seg, which progressively predicts the next-scale mask by explicitly modeling dependencies across all previous scales within a unified architecture. AR-Seg introduces three innovations: (1) a multi-scale mask autoencoder that quantizes the mask into multi-scale token maps to capture hierarchical anatomical structures, (2) a next-scale autoregressive mechanism that progressively predicts next-scale masks to enable sufficient inter-scale dependencies, and (3) a consensus-aggregation strategy that combines multiple sampled results to generate a more accurate mask, further improving segmentation robustness. Extensive experimental results on two benchmark datasets with different modalities demonstrate that AR-Seg outperforms state-of-the-art methods while explicitly visualizing the intermediate coarse-to-fine segmentation process.
HiDiff: Hybrid Diffusion Framework for Medical Image Segmentation
Jul 03, 2024

Abstract:Medical image segmentation has been significantly advanced with the rapid development of deep learning (DL) techniques. Existing DL-based segmentation models are typically discriminative; i.e., they aim to learn a mapping from the input image to segmentation masks. However, these discriminative methods neglect the underlying data distribution and intrinsic class characteristics, suffering from unstable feature space. In this work, we propose to complement discriminative segmentation methods with the knowledge of underlying data distribution from generative models. To that end, we propose a novel hybrid diffusion framework for medical image segmentation, termed HiDiff, which can synergize the strengths of existing discriminative segmentation models and new generative diffusion models. HiDiff comprises two key components: discriminative segmentor and diffusion refiner. First, we utilize any conventional trained segmentation models as discriminative segmentor, which can provide a segmentation mask prior for diffusion refiner. Second, we propose a novel binary Bernoulli diffusion model (BBDM) as the diffusion refiner, which can effectively, efficiently, and interactively refine the segmentation mask by modeling the underlying data distribution. Third, we train the segmentor and BBDM in an alternate-collaborative manner to mutually boost each other. Extensive experimental results on abdomen organ, brain tumor, polyps, and retinal vessels segmentation datasets, covering four widely-used modalities, demonstrate the superior performance of HiDiff over existing medical segmentation algorithms, including the state-of-the-art transformer- and diffusion-based ones. In addition, HiDiff excels at segmenting small objects and generalizing to new datasets. Source codes are made available at https://github.com/takimailto/HiDiff.
FLDM-VTON: Faithful Latent Diffusion Model for Virtual Try-on
Apr 22, 2024Abstract:Despite their impressive generative performance, latent diffusion model-based virtual try-on (VTON) methods lack faithfulness to crucial details of the clothes, such as style, pattern, and text. To alleviate these issues caused by the diffusion stochastic nature and latent supervision, we propose a novel Faithful Latent Diffusion Model for VTON, termed FLDM-VTON. FLDM-VTON improves the conventional latent diffusion process in three major aspects. First, we propose incorporating warped clothes as both the starting point and local condition, supplying the model with faithful clothes priors. Second, we introduce a novel clothes flattening network to constrain generated try-on images, providing clothes-consistent faithful supervision. Third, we devise a clothes-posterior sampling for faithful inference, further enhancing the model performance over conventional clothes-agnostic Gaussian sampling. Extensive experimental results on the benchmark VITON-HD and Dress Code datasets demonstrate that our FLDM-VTON outperforms state-of-the-art baselines and is able to generate photo-realistic try-on images with faithful clothing details.
Low-dose CT Denoising with Language-engaged Dual-space Alignment
Mar 10, 2024



Abstract:While various deep learning methods were proposed for low-dose computed tomography (CT) denoising, they often suffer from over-smoothing, blurring, and lack of explainability. To alleviate these issues, we propose a plug-and-play Language-Engaged Dual-space Alignment loss (LEDA) to optimize low-dose CT denoising models. Our idea is to leverage large language models (LLMs) to align denoised CT and normal dose CT images in both the continuous perceptual space and discrete semantic space, which is the first LLM-based scheme for low-dose CT denoising. LEDA involves two steps: the first is to pretrain an LLM-guided CT autoencoder, which can encode a CT image into continuous high-level features and quantize them into a token space to produce semantic tokens derived from the LLM's vocabulary; and the second is to minimize the discrepancy between the denoised CT images and normal dose CT in terms of both encoded high-level features and quantized token embeddings derived by the LLM-guided CT autoencoder. Extensive experimental results on two public LDCT denoising datasets demonstrate that our LEDA can enhance existing denoising models in terms of quantitative metrics and qualitative evaluation, and also provide explainability through language-level image understanding. Source code is available at https://github.com/hao1635/LEDA.
HOPE: Hybrid-granularity Ordinal Prototype Learning for Progression Prediction of Mild Cognitive Impairment
Jan 19, 2024



Abstract:Mild cognitive impairment (MCI) is often at high risk of progression to Alzheimer's disease (AD). Existing works to identify the progressive MCI (pMCI) typically require MCI subtype labels, pMCI vs. stable MCI (sMCI), determined by whether or not an MCI patient will progress to AD after a long follow-up. However, prospectively acquiring MCI subtype data is time-consuming and resource-intensive; the resultant small datasets could lead to severe overfitting and difficulty in extracting discriminative information. Inspired by that various longitudinal biomarkers and cognitive measurements present an ordinal pathway on AD progression, we propose a novel Hybrid-granularity Ordinal PrototypE learning (HOPE) method to characterize AD ordinal progression for MCI progression prediction. First, HOPE learns an ordinal metric space that enables progression prediction by prototype comparison. Second, HOPE leverages a novel hybrid-granularity ordinal loss to learn the ordinal nature of AD via effectively integrating instance-to-instance ordinality, instance-to-class compactness, and class-to-class separation. Third, to make the prototype learning more stable, HOPE employs an exponential moving average strategy to learn the global prototypes of NC and AD dynamically. Experimental results on the internal ADNI and the external NACC datasets demonstrate the superiority of the proposed HOPE over existing state-of-the-art methods as well as its interpretability. Source code is made available at https://github.com/thibault-wch/HOPE-for-mild-cognitive-impairment.
* IEEE Journal of Biomedical and Health Informatics, 2024
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge