"cancer detection": models, code, and papers
Efficient High-Resolution Deep Learning: A Survey
Jul 26, 2022
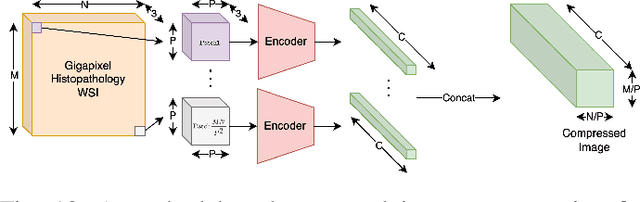

Cameras in modern devices such as smartphones, satellites and medical equipment are capable of capturing very high resolution images and videos. Such high-resolution data often need to be processed by deep learning models for cancer detection, automated road navigation, weather prediction, surveillance, optimizing agricultural processes and many other applications. Using high-resolution images and videos as direct inputs for deep learning models creates many challenges due to their high number of parameters, computation cost, inference latency and GPU memory consumption. Simple approaches such as resizing the images to a lower resolution are common in the literature, however, they typically significantly decrease accuracy. Several works in the literature propose better alternatives in order to deal with the challenges of high-resolution data and improve accuracy and speed while complying with hardware limitations and time restrictions. This survey describes such efficient high-resolution deep learning methods, summarizes real-world applications of high-resolution deep learning, and provides comprehensive information about available high-resolution datasets.
Meta-information-aware Dual-path Transformer for Differential Diagnosis of Multi-type Pancreatic Lesions in Multi-phase CT
Mar 02, 2023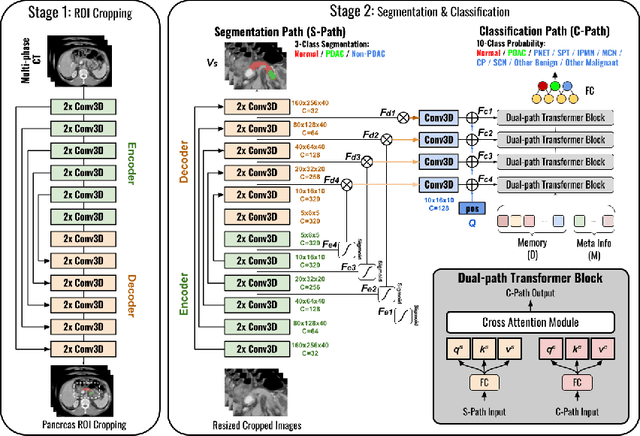
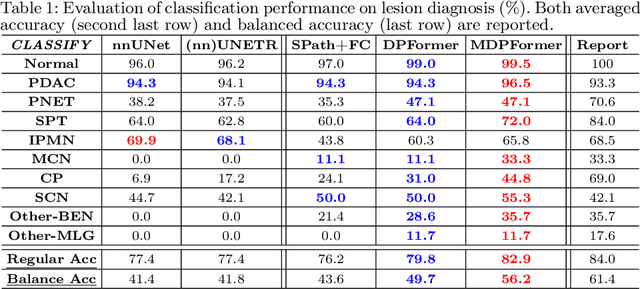
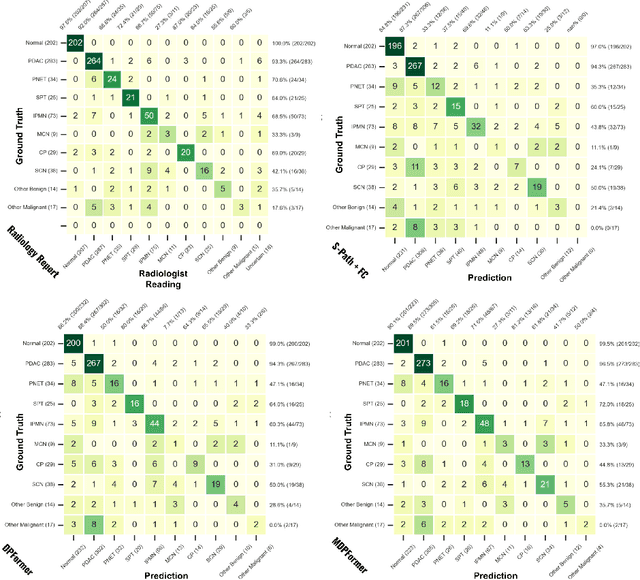
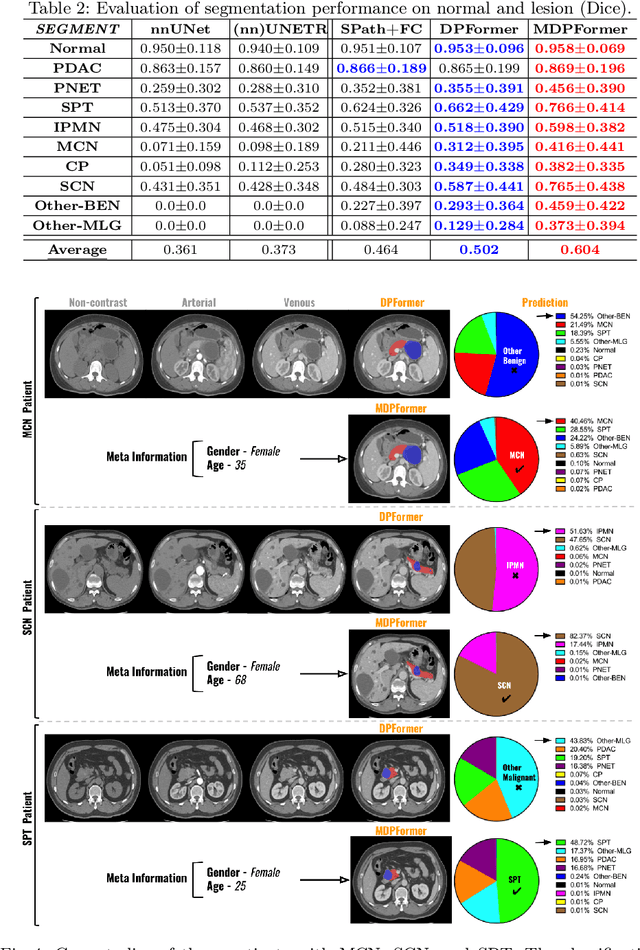
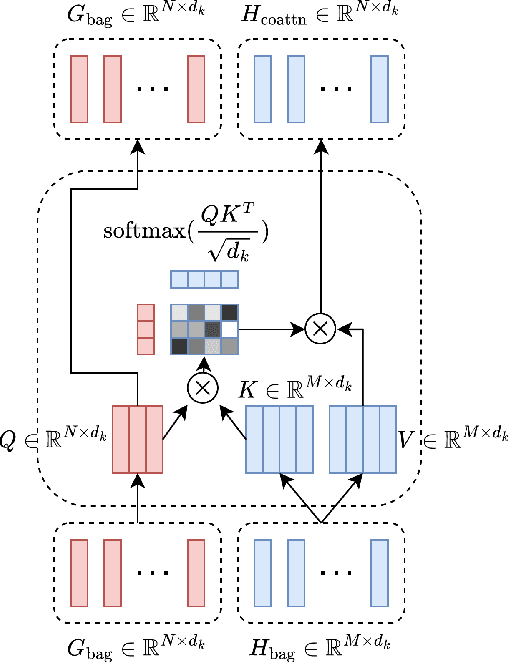
Pancreatic cancer is one of the leading causes of cancer-related death. Accurate detection, segmentation, and differential diagnosis of the full taxonomy of pancreatic lesions, i.e., normal, seven major types of lesions, and other lesions, is critical to aid the clinical decision-making of patient management and treatment. However, existing works focus on segmentation and classification for very specific lesion types (PDAC) or groups. Moreover, none of the previous work considers using lesion prevalence-related non-imaging patient information to assist the differential diagnosis. To this end, we develop a meta-information-aware dual-path transformer and exploit the feasibility of classification and segmentation of the full taxonomy of pancreatic lesions. Specifically, the proposed method consists of a CNN-based segmentation path (S-path) and a transformer-based classification path (C-path). The S-path focuses on initial feature extraction by semantic segmentation using a UNet-based network. The C-path utilizes both the extracted features and meta-information for patient-level classification based on stacks of dual-path transformer blocks that enhance the modeling of global contextual information. A large-scale multi-phase CT dataset of 3,096 patients with pathology-confirmed pancreatic lesion class labels, voxel-wise manual annotations of lesions from radiologists, and patient meta-information, was collected for training and evaluations. Our results show that our method can enable accurate classification and segmentation of the full taxonomy of pancreatic lesions, approaching the accuracy of the radiologist's report and significantly outperforming previous baselines. Results also show that adding the common meta-information, i.e., gender and age, can boost the model's performance, thus demonstrating the importance of meta-information for aiding pancreatic disease diagnosis.
Deep Semi-supervised Metric Learning with Dual Alignment for Cervical Cancer Cell Detection
Apr 07, 2021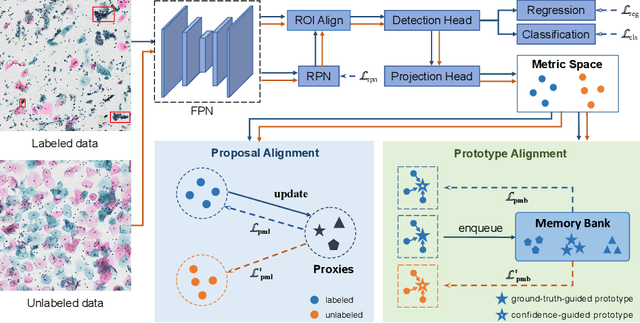
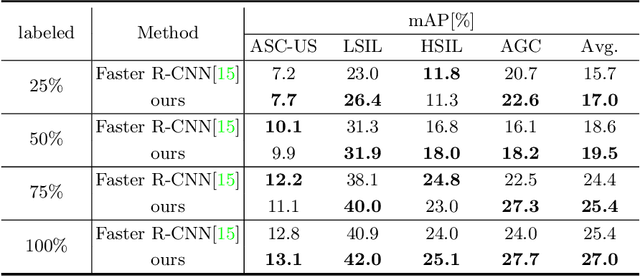
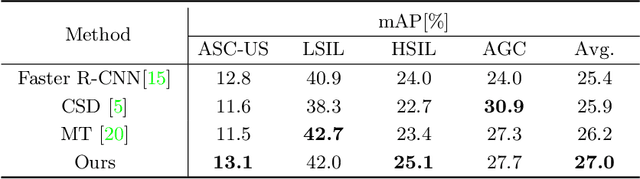
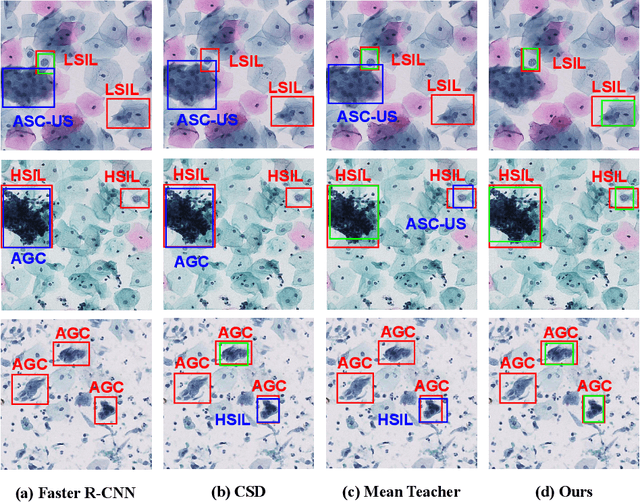
With availability of huge amounts of labeled data, deep learning has achieved unprecedented success in various object detection tasks. However, large-scale annotations for medical images are extremely challenging to be acquired due to the high demand of labour and expertise. To address this difficult issue, in this paper we propose a novel semi-supervised deep metric learning method to effectively leverage both labeled and unlabeled data with application to cervical cancer cell detection. Different from previous methods, our model learns an embedding metric space and conducts dual alignment of semantic features on both the proposal and prototype levels. First, on the proposal level, we generate pseudo labels for the unlabeled data to align the proposal features with learnable class proxies derived from the labeled data. Furthermore, we align the prototypes generated from each mini-batch of labeled and unlabeled data to alleviate the influence of possibly noisy pseudo labels. Moreover, we adopt a memory bank to store the labeled prototypes and hence significantly enrich the metric learning information from larger batches. To comprehensively validate the method, we construct a large-scale dataset for semi-supervised cervical cancer cell detection for the first time, consisting of 240,860 cervical cell images in total. Extensive experiments show our proposed method outperforms other state-of-the-art semi-supervised approaches consistently, demonstrating efficacy of deep semi-supervised metric learning with dual alignment on improving cervical cancer cell detection performance.
Extraction of Pulmonary Airway in CT Scans Using Deep Fully Convolutional Networks
Aug 12, 2022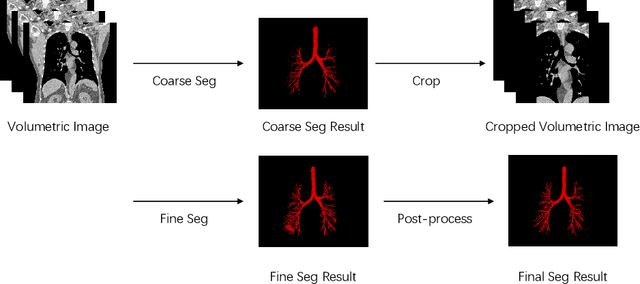
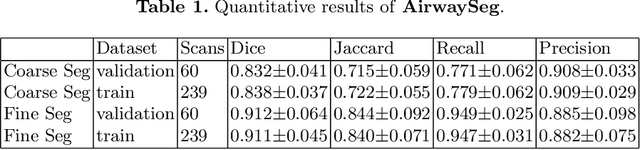

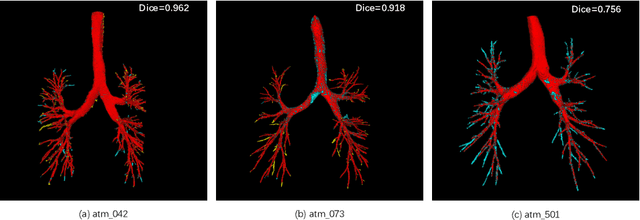
Accurate, automatic and complete extraction of pulmonary airway in medical images plays an important role in analyzing thoracic CT volumes such as lung cancer detection, chronic obstructive pulmonary disease (COPD), and bronchoscopic-assisted surgery navigation. However, this task remains challenges, due to the complex tree-like structure of the airways. In this technical report, we use two-stage fully convolutional networks (FCNs) to automatically segment pulmonary airway in thoracic CT scans from multi-sites. Specifically, we firstly adopt a 3D FCN with U-shape network architecture to segment pulmonary airway in a coarse resolution in order to accelerate medical image analysis pipeline. And then another one 3D FCN is trained to segment pulmonary airway in a fine resolution. In the 2022 MICCAI Multi-site Multi-domain Airway Tree Modeling (ATM) Challenge, the reported method was evaluated on the public training set of 300 cases and independent private validation set of 50 cases. The resulting Dice Similarity Coefficient (DSC) is 0.914 $\pm$ 0.040, False Negative Error (FNE) is 0.079 $\pm$ 0.042, and False Positive Error (FPE) is 0.090 $\pm$ 0.066 on independent private validation set.
Gene selection from microarray expression data: A Multi-objective PSO with adaptive K-nearest neighborhood
May 27, 2022
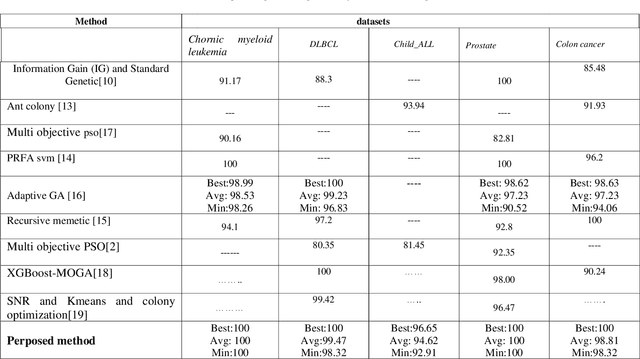
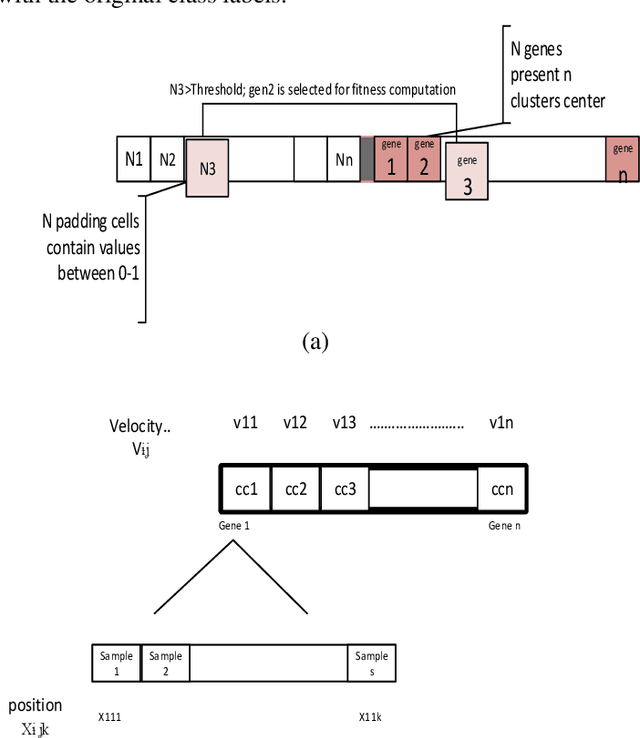
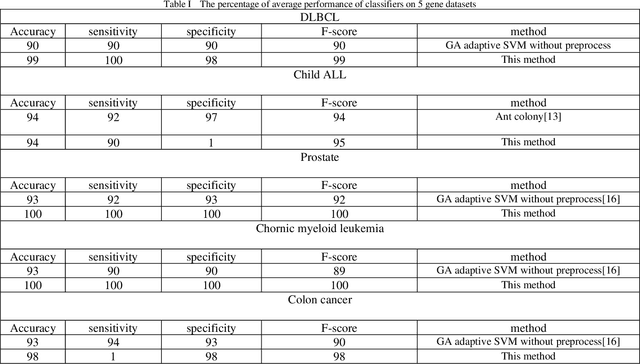
Cancer detection is one of the key research topics in the medical field. Accurate detection of different cancer types is valuable in providing better treatment facilities and risk minimization for patients. This paper deals with the classification problem of human cancer diseases by using gene expression data. It is presented a new methodology to analyze microarray datasets and efficiently classify cancer diseases. The new method first employs Signal to Noise Ratio (SNR) to find a list of a small subset of non-redundant genes. Then, after normalization, it is used Multi-Objective Particle Swarm Optimization (MOPSO) for feature selection and employed Adaptive K-Nearest Neighborhood (KNN) for cancer disease classification. This method improves the classification accuracy of cancer classification by reducing the number of features. The proposed methodology is evaluated by classifying cancer diseases in five cancer datasets. The results are compared with the most recent approaches, which increases the classification accuracy in each dataset.
Autofluorescence Bronchoscopy Video Analysis for Lesion Frame Detection
Mar 21, 2023
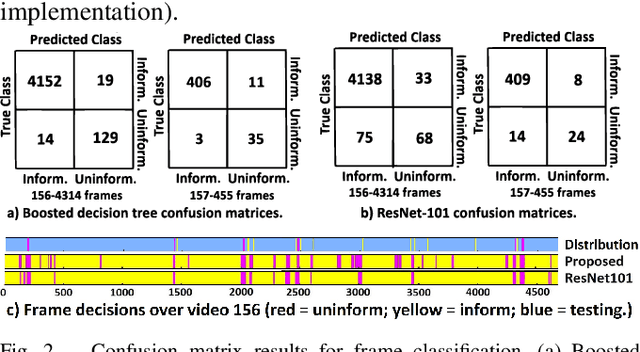
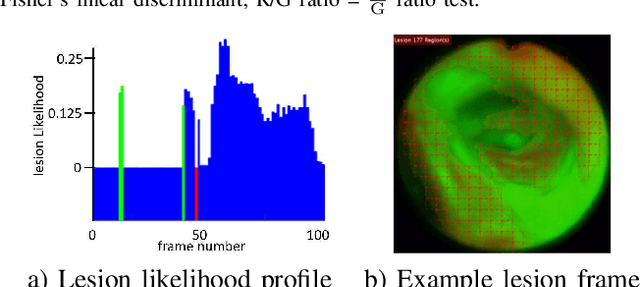
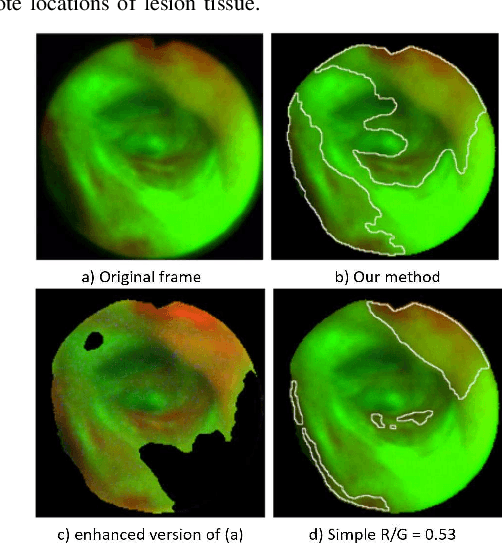
Because of the significance of bronchial lesions as indicators of early lung cancer and squamous cell carcinoma, a critical need exists for early detection of bronchial lesions. Autofluorescence bronchoscopy (AFB) is a primary modality used for bronchial lesion detection, as it shows high sensitivity to suspicious lesions. The physician, however, must interactively browse a long video stream to locate lesions, making the search exceedingly tedious and error prone. Unfortunately, limited research has explored the use of automated AFB video analysis for efficient lesion detection. We propose a robust automatic AFB analysis approach that distinguishes informative and uninformative AFB video frames in a video. In addition, for the informative frames, we determine the frames containing potential lesions and delineate candidate lesion regions. Our approach draws upon a combination of computer-based image analysis, machine learning, and deep learning. Thus, the analysis of an AFB video stream becomes more tractable. Tests with patient AFB video indicate that $\ge$97\% of frames were correctly labeled as informative or uninformative. In addition, $\ge$97\% of lesion frames were correctly identified, with false positive and false negative rates $\le$3\%.
Improving the diagnosis of breast cancer based on biophysical ultrasound features utilizing machine learning
Jul 13, 2022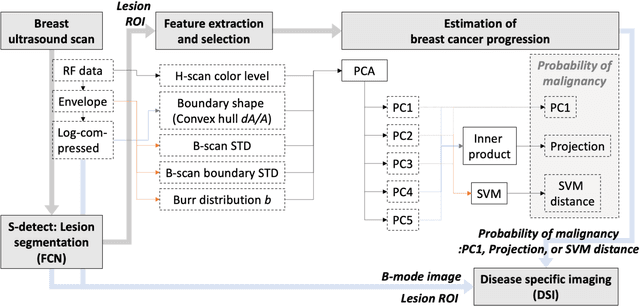

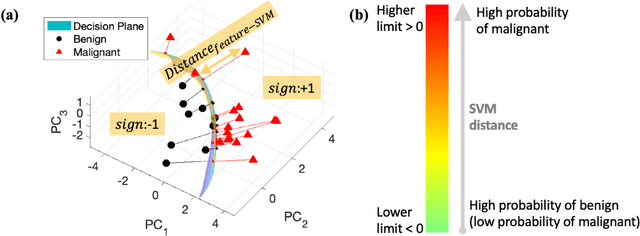
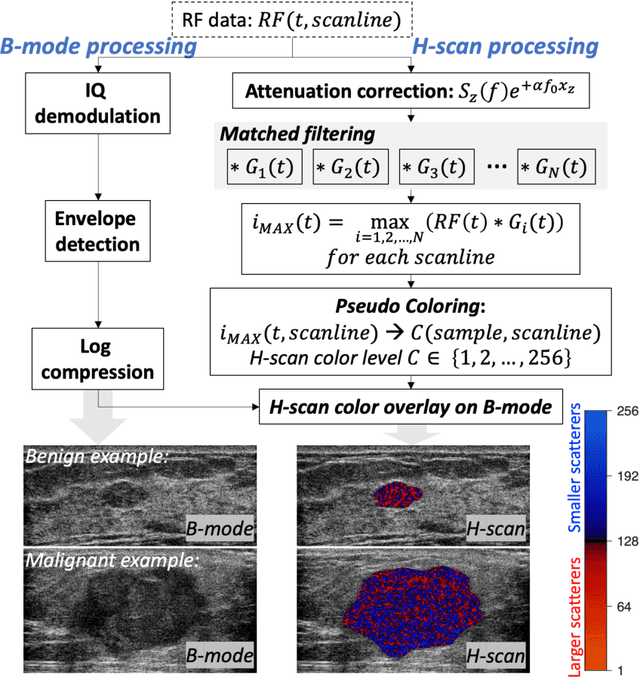
The improved diagnostic accuracy of ultrasound breast examinations remains an important goal. In this study, we propose a biophysical feature based machine learning method for breast cancer detection to improve the performance beyond a benchmark deep learning algorithm and to furthermore provide a color overlay visual map of the probability of malignancy within a lesion. This overall framework is termed disease specific imaging. Previously, 150 breast lesions were segmented and classified utilizing a modified fully convolutional network and a modified GoogLeNet, respectively. In this study multiparametric analysis was performed within the contoured lesions. Features were extracted from ultrasound radiofrequency, envelope, and log compressed data based on biophysical and morphological models. The support vector machine with a Gaussian kernel constructed a nonlinear hyperplane, and we calculated the distance between the hyperplane and data point of each feature in multiparametric space. The distance can quantitatively assess a lesion, and suggest the probability of malignancy that is color coded and overlaid onto B mode images. Training and evaluation were performed on in vivo patient data. The overall accuracy for the most common types and sizes of breast lesions in our study exceeded 98.0% for classification and 0.98 for an area under the receiver operating characteristic curve, which is more precise than the performance of radiologists and a deep learning system. Further, the correlation between the probability and BI RADS enables a quantitative guideline to predict breast cancer. Therefore, we anticipate that the proposed framework can help radiologists achieve more accurate and convenient breast cancer classification and detection.
Feasibility of Colon Cancer Detection in Confocal Laser Microscopy Images Using Convolution Neural Networks
Dec 05, 2018
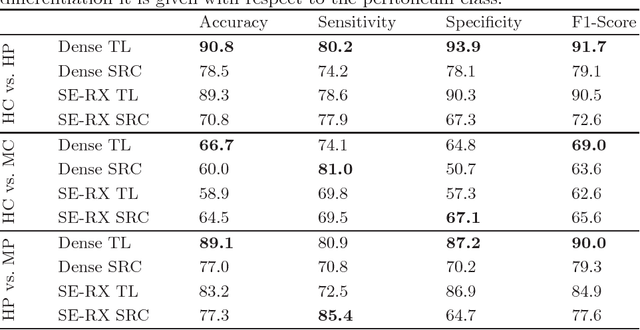
Histological evaluation of tissue samples is a typical approach to identify colorectal cancer metastases in the peritoneum. For immediate assessment, reliable and real-time in-vivo imaging would be required. For example, intraoperative confocal laser microscopy has been shown to be suitable for distinguishing organs and also malignant and benign tissue. So far, the analysis is done by human experts. We investigate the feasibility of automatic colon cancer classification from confocal laser microscopy images using deep learning models. We overcome very small dataset sizes through transfer learning with state-of-the-art architectures. We achieve an accuracy of 89.1% for cancer detection in the peritoneum which indicates viability as an intraoperative decision support system.
Classification of Breast Tumours Based on Histopathology Images Using Deep Features and Ensemble of Gradient Boosting Methods
Sep 03, 2022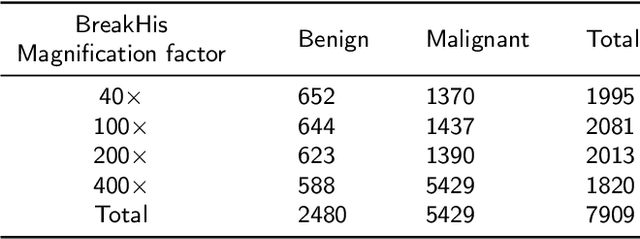
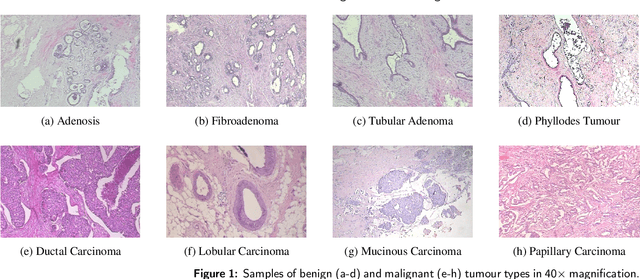
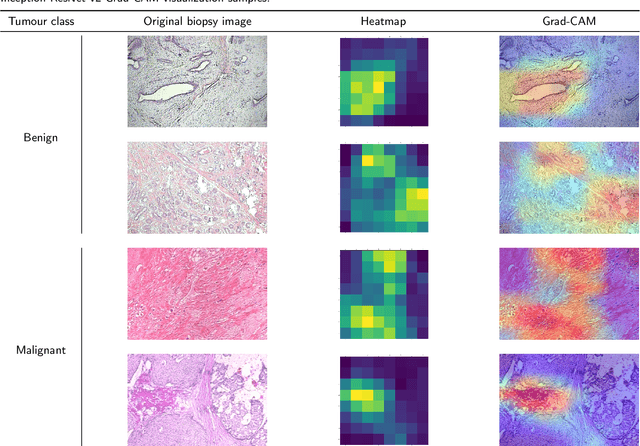
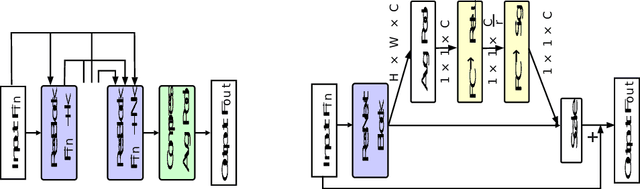
Breast cancer is the most common cancer among women worldwide. Early-stage diagnosis of breast cancer can significantly improve the efficiency of treatment. Computer-aided diagnosis (CAD) systems are widely adopted in this issue due to their reliability, accuracy and affordability. There are different imaging techniques for a breast cancer diagnosis; one of the most accurate ones is histopathology which is used in this paper. Deep feature transfer learning is used as the main idea of the proposed CAD system's feature extractor. Although 16 different pre-trained networks have been tested in this study, our main focus is on the classification phase. The Inception-ResNet-v2 which has both residual and inception networks profits together has shown the best feature extraction capability in the case of breast cancer histopathology images among all tested CNNs. In the classification phase, the ensemble of CatBoost, XGBoost and LightGBM has provided the best average accuracy. The BreakHis dataset was used to evaluate the proposed method. BreakHis contains 7909 histopathology images (2,480 benign and 5,429 malignant) in four magnification factors. The proposed method's accuracy (IRv2-CXL) using 70% of BreakHis dataset as training data in 40x, 100x, 200x and 400x magnification is 96.82%, 95.84%, 97.01% and 96.15%, respectively. Most studies on automated breast cancer detection have focused on feature extraction, which made us attend to the classification phase. IRv2-CXL has shown better or comparable results in all magnifications due to using the soft voting ensemble method which could combine the advantages of CatBoost, XGBoost and LightGBM together.
EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks
Dec 06, 2022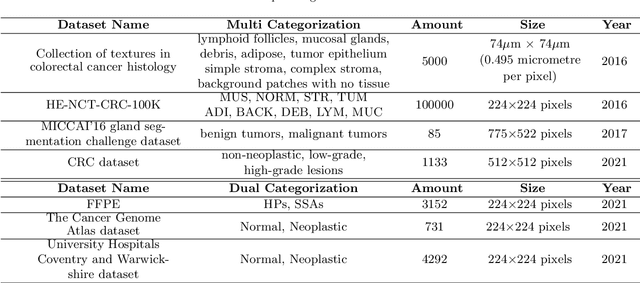
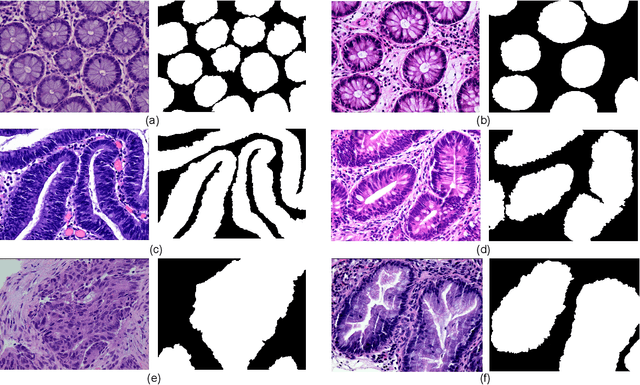
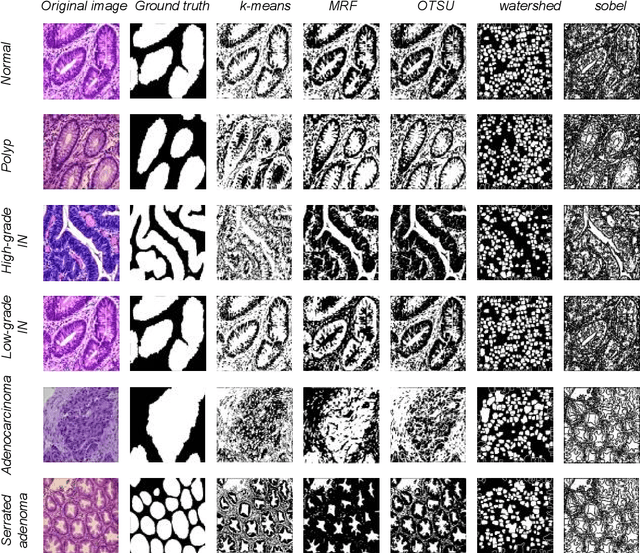
Background and Purpose: Colorectal cancer is a common fatal malignancy, the fourth most common cancer in men, and the third most common cancer in women worldwide. Timely detection of cancer in its early stages is essential for treating the disease. Currently, there is a lack of datasets for histopathological image segmentation of rectal cancer, which often hampers the assessment accuracy when computer technology is used to aid in diagnosis. Methods: This present study provided a new publicly available Enteroscope Biopsy Histopathological Hematoxylin and Eosin Image Dataset for Image Segmentation Tasks (EBHI-Seg). To demonstrate the validity and extensiveness of EBHI-Seg, the experimental results for EBHI-Seg are evaluated using classical machine learning methods and deep learning methods. Results: The experimental results showed that deep learning methods had a better image segmentation performance when utilizing EBHI-Seg. The maximum accuracy of the Dice evaluation metric for the classical machine learning method is 0.948, while the Dice evaluation metric for the deep learning method is 0.965. Conclusion: This publicly available dataset contained 5,170 images of six types of tumor differentiation stages and the corresponding ground truth images. The dataset can provide researchers with new segmentation algorithms for medical diagnosis of colorectal cancer, which can be used in the clinical setting to help doctors and patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge