Deformable Medical Image Registration
Papers and Code
Attention-Driven Framework for Non-Rigid Medical Image Registration
Feb 06, 2026Deformable medical image registration is a fundamental task in medical image analysis with applications in disease diagnosis, treatment planning, and image-guided interventions. Despite significant advances in deep learning based registration methods, accurately aligning images with large deformations while preserving anatomical plausibility remains a challenging task. In this paper, we propose a novel Attention-Driven Framework for Non-Rigid Medical Image Registration (AD-RegNet) that employs attention mechanisms to guide the registration process. Our approach combines a 3D UNet backbone with bidirectional cross-attention, which establishes correspondences between moving and fixed images at multiple scales. We introduce a regional adaptive attention mechanism that focuses on anatomically relevant structures, along with a multi-resolution deformation field synthesis approach for accurate alignment. The method is evaluated on two distinct datasets: DIRLab for thoracic 4D CT scans and IXI for brain MRI scans, demonstrating its versatility across different anatomical structures and imaging modalities. Experimental results demonstrate that our approach achieves performance competitive with state-of-the-art methods on the IXI and DIRLab datasets. The proposed method maintains a favorable balance between registration accuracy and computational efficiency, making it suitable for clinical applications. A comprehensive evaluation using normalized cross-correlation (NCC), mean squared error (MSE), structural similarity (SSIM), Jacobian determinant, and target registration error (TRE) indicates that attention-guided registration improves alignment accuracy while ensuring anatomically plausible deformations.
Unsupervised MRI-US Multimodal Image Registration with Multilevel Correlation Pyramidal Optimization
Feb 06, 2026Surgical navigation based on multimodal image registration has played a significant role in providing intraoperative guidance to surgeons by showing the relative position of the target area to critical anatomical structures during surgery. However, due to the differences between multimodal images and intraoperative image deformation caused by tissue displacement and removal during the surgery, effective registration of preoperative and intraoperative multimodal images faces significant challenges. To address the multimodal image registration challenges in Learn2Reg 2025, an unsupervised multimodal medical image registration method based on multilevel correlation pyramidal optimization (MCPO) is designed to solve these problems. First, the features of each modality are extracted based on the modality independent neighborhood descriptor, and the multimodal images is mapped to the feature space. Second, a multilevel pyramidal fusion optimization mechanism is designed to achieve global optimization and local detail complementation of the displacement field through dense correlation analysis and weight-balanced coupled convex optimization for input features at different scales. Our method focuses on the ReMIND2Reg task in Learn2Reg 2025. Based on the results, our method achieved the first place in the validation phase and test phase of ReMIND2Reg. The MCPO is also validated on the Resect dataset, achieving an average TRE of 1.798 mm. This demonstrates the broad applicability of our method in preoperative-to-intraoperative image registration. The code is avaliable at https://github.com/wjiazheng/MCPO.
LDRNet: Large Deformation Registration Model for Chest CT Registration
Feb 02, 2026Most of the deep learning based medical image registration algorithms focus on brain image registration tasks.Compared with brain registration, the chest CT registration has larger deformation, more complex background and region over-lap. In this paper, we propose a fast unsupervised deep learning method, LDRNet, for large deformation image registration of chest CT images. We first predict a coarse resolution registration field, then refine it from coarse to fine. We propose two innovative technical components: 1) a refine block that is used to refine the registration field in different resolutions, 2) a rigid block that is used to learn transformation matrix from high-level features. We train and evaluate our model on the private dataset and public dataset SegTHOR. We compare our performance with state-of-the-art traditional registration methods as well as deep learning registration models VoxelMorph, RCN, and LapIRN. The results demonstrate that our model achieves state-of-the-art performance for large deformation images registration and is much faster.
Interpretable Unsupervised Deformable Image Registration via Confidence-bound Multi-Hop Visual Reasoning
Jan 30, 2026Unsupervised deformable image registration requires aligning complex anatomical structures without reference labels, making interpretability and reliability critical. Existing deep learning methods achieve considerable accuracy but often lack transparency, leading to error drift and reduced clinical trust. We propose a novel Multi-Hop Visual Chain of Reasoning (VCoR) framework that reformulates registration as a progressive reasoning process. Inspired by the iterative nature of clinical decision-making, each visual reasoning hop integrates a Localized Spatial Refinement (LSR) module to enrich feature representations and a Cross-Reference Attention (CRA) mechanism that leads the iterative refinement process, preserving anatomical consistency. This multi-hop strategy enables robust handling of large deformations and produces a transparent sequence of intermediate predictions with a theoretical bound. Beyond accuracy, our framework offers built-in interpretability by estimating uncertainty via the stability and convergence of deformation fields across hops. Extensive evaluations on two challenging public datasets, DIR-Lab 4D CT (lung) and IXI T1-weighted MRI (brain), demonstrate that VCoR achieves competitive registration accuracy while offering rich intermediate visualizations and confidence measures. By embedding an implicit visual reasoning paradigm, we present an interpretable, reliable, and clinically viable unsupervised medical image registration.
Advanced Geometric Correction Algorithms for 3D Medical Reconstruction: Comparison of Computed Tomography and Macroscopic Imaging
Jan 30, 2026This paper introduces a hybrid two-stage registration framework for reconstructing three-dimensional (3D) kidney anatomy from macroscopic slices, using CT-derived models as the geometric reference standard. The approach addresses the data-scarcity and high-distortion challenges typical of macroscopic imaging, where fully learning-based registration (e.g., VoxelMorph) often fails to generalize due to limited training diversity and large nonrigid deformations that exceed the capture range of unconstrained convolutional filters. In the proposed pipeline, the Optimal Cross-section Matching (OCM) algorithm first performs constrained global alignment: translation, rotation, and uniform scaling to establish anatomically consistent slice initialization. Next, a lightweight deep-learning refinement network, inspired by VoxelMorph, predicts residual local deformations between consecutive slices. The core novelty of this architecture lies in its hierarchical decomposition of the registration manifold. This hybrid OCM+DL design integrates explicit geometric priors with the flexible learning capacity of neural networks, ensuring stable optimization and plausible deformation fields even with few training examples. Experiments on an original dataset of 40 kidneys demonstrated better results compared to single-stage baselines. The pipeline maintains physical calibration via Hough-based grid detection and employs Bezier-based contour smoothing for robust meshing and volume estimation. Although validated on kidney data, the proposed framework generalizes to other soft-tissue organs reconstructed from optical or photographic cross-sections. By decoupling interpretable global optimization from data-efficient deep refinement, the method advances the precision, reproducibility, and anatomical realism of multimodal 3D reconstructions for surgical planning, morphological assessment, and medical education.
Dynamic Stream Network for Combinatorial Explosion Problem in Deformable Medical Image Registration
Dec 22, 2025



Combinatorial explosion problem caused by dual inputs presents a critical challenge in Deformable Medical Image Registration (DMIR). Since DMIR processes two images simultaneously as input, the combination relationships between features has grown exponentially, ultimately the model considers more interfering features during the feature modeling process. Introducing dynamics in the receptive fields and weights of the network enable the model to eliminate the interfering features combination and model the potential feature combination relationships. In this paper, we propose the Dynamic Stream Network (DySNet), which enables the receptive fields and weights to be dynamically adjusted. This ultimately enables the model to ignore interfering feature combinations and model the potential feature relationships. With two key innovations: 1) Adaptive Stream Basin (AdSB) module dynamically adjusts the shape of the receptive field, thereby enabling the model to focus on the feature relationships with greater correlation. 2) Dynamic Stream Attention (DySA) mechanism generates dynamic weights to search for more valuable feature relationships. Extensive experiments have shown that DySNet consistently outperforms the most advanced DMIR methods, highlighting its outstanding generalization ability. Our code will be released on the website: https://github.com/ShaochenBi/DySNet.
Test Time Optimized Generalized AI-based Medical Image Registration Method
Dec 16, 2025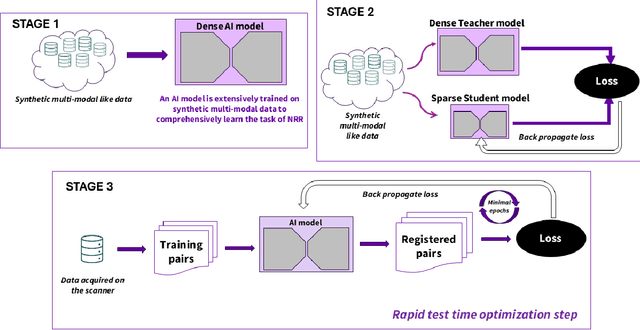
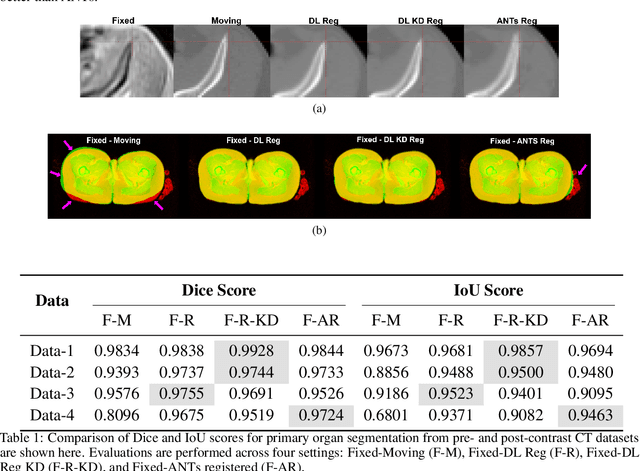
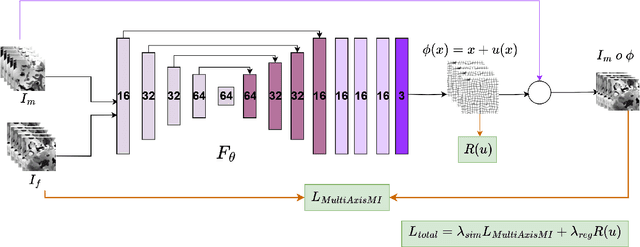
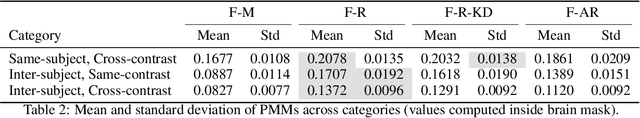
Medical image registration is critical for aligning anatomical structures across imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound. Among existing techniques, non-rigid registration (NRR) is particularly challenging due to the need to capture complex anatomical deformations caused by physiological processes like respiration or contrast-induced signal variations. Traditional NRR methods, while theoretically robust, often require extensive parameter tuning and incur high computational costs, limiting their use in real-time clinical workflows. Recent deep learning (DL)-based approaches have shown promise; however, their dependence on task-specific retraining restricts scalability and adaptability in practice. These limitations underscore the need for efficient, generalizable registration frameworks capable of handling heterogeneous imaging contexts. In this work, we introduce a novel AI-driven framework for 3D non-rigid registration that generalizes across multiple imaging modalities and anatomical regions. Unlike conventional methods that rely on application-specific models, our approach eliminates anatomy- or modality-specific customization, enabling streamlined integration into diverse clinical environments.
Efficient Large-Deformation Medical Image Registration via Recurrent Dynamic Correlation
Oct 25, 2025Deformable image registration estimates voxel-wise correspondences between images through spatial transformations, and plays a key role in medical imaging. While deep learning methods have significantly reduced runtime, efficiently handling large deformations remains a challenging task. Convolutional networks aggregate local features but lack direct modeling of voxel correspondences, promoting recent works to explore explicit feature matching. Among them, voxel-to-region matching is more efficient for direct correspondence modeling by computing local correlation features whithin neighbourhoods, while region-to-region matching incurs higher redundancy due to excessive correlation pairs across large regions. However, the inherent locality of voxel-to-region matching hinders the capture of long-range correspondences required for large deformations. To address this, we propose a Recurrent Correlation-based framework that dynamically relocates the matching region toward more promising positions. At each step, local matching is performed with low cost, and the estimated offset guides the next search region, supporting efficient convergence toward large deformations. In addition, we uses a lightweight recurrent update module with memory capacity and decouples motion-related and texture features to suppress semantic redundancy. We conduct extensive experiments on brain MRI and abdominal CT datasets under two settings: with and without affine pre-registration. Results show that our method exibits a strong accuracy-computation trade-off, surpassing or matching the state-of-the-art performance. For example, it achieves comparable performance on the non-affine OASIS dataset, while using only 9.5% of the FLOPs and running 96% faster than RDP, a representative high-performing method.
Revisiting Data Scaling Law for Medical Segmentation
Nov 17, 2025The population loss of trained deep neural networks often exhibits power law scaling with the size of the training dataset, guiding significant performance advancements in deep learning applications. In this study, we focus on the scaling relationship with data size in the context of medical anatomical segmentation, a domain that remains underexplored. We analyze scaling laws for anatomical segmentation across 15 semantic tasks and 4 imaging modalities, demonstrating that larger datasets significantly improve segmentation performance, following similar scaling trends. Motivated by the topological isomorphism in images sharing anatomical structures, we evaluate the impact of deformation-guided augmentation strategies on data scaling laws, specifically random elastic deformation and registration-guided deformation. We also propose a novel, scalable image augmentation approach that generates diffeomorphic mappings from geodesic subspace based on image registration to introduce realistic deformation. Our experimental results demonstrate that both registered and generated deformation-based augmentation considerably enhance data utilization efficiency. The proposed generated deformation method notably achieves superior performance and accelerated convergence, surpassing standard power law scaling trends without requiring additional data. Overall, this work provides insights into the understanding of segmentation scalability and topological variation impact in medical imaging, thereby leading to more efficient model development with reduced annotation and computational costs.
AtlasMorph: Learning conditional deformable templates for brain MRI
Nov 17, 2025



Deformable templates, or atlases, are images that represent a prototypical anatomy for a population, and are often enhanced with probabilistic anatomical label maps. They are commonly used in medical image analysis for population studies and computational anatomy tasks such as registration and segmentation. Because developing a template is a computationally expensive process, relatively few templates are available. As a result, analysis is often conducted with sub-optimal templates that are not truly representative of the study population, especially when there are large variations within this population. We propose a machine learning framework that uses convolutional registration neural networks to efficiently learn a function that outputs templates conditioned on subject-specific attributes, such as age and sex. We also leverage segmentations, when available, to produce anatomical segmentation maps for the resulting templates. The learned network can also be used to register subject images to the templates. We demonstrate our method on a compilation of 3D brain MRI datasets, and show that it can learn high-quality templates that are representative of populations. We find that annotated conditional templates enable better registration than their unlabeled unconditional counterparts, and outperform other templates construction methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge