S. Mazdak Abulnaga
Unified Brain Surface and Volume Registration
Dec 22, 2025Abstract:Accurate registration of brain MRI scans is fundamental for cross-subject analysis in neuroscientific studies. This involves aligning both the cortical surface of the brain and the interior volume. Traditional methods treat volumetric and surface-based registration separately, which often leads to inconsistencies that limit downstream analyses. We propose a deep learning framework, NeurAlign, that registers $3$D brain MRI images by jointly aligning both cortical and subcortical regions through a unified volume-and-surface-based representation. Our approach leverages an intermediate spherical coordinate space to bridge anatomical surface topology with volumetric anatomy, enabling consistent and anatomically accurate alignment. By integrating spherical registration into the learning, our method ensures geometric coherence between volume and surface domains. In a series of experiments on both in-domain and out-of-domain datasets, our method consistently outperforms both classical and machine learning-based registration methods -- improving the Dice score by up to 7 points while maintaining regular deformation fields. Additionally, it is orders of magnitude faster than the standard method for this task, and is simpler to use because it requires no additional inputs beyond an MRI scan. With its superior accuracy, fast inference, and ease of use, NeurAlign sets a new standard for joint cortical and subcortical registration.
AtlasMorph: Learning conditional deformable templates for brain MRI
Nov 17, 2025



Abstract:Deformable templates, or atlases, are images that represent a prototypical anatomy for a population, and are often enhanced with probabilistic anatomical label maps. They are commonly used in medical image analysis for population studies and computational anatomy tasks such as registration and segmentation. Because developing a template is a computationally expensive process, relatively few templates are available. As a result, analysis is often conducted with sub-optimal templates that are not truly representative of the study population, especially when there are large variations within this population. We propose a machine learning framework that uses convolutional registration neural networks to efficiently learn a function that outputs templates conditioned on subject-specific attributes, such as age and sex. We also leverage segmentations, when available, to produce anatomical segmentation maps for the resulting templates. The learned network can also be used to register subject images to the templates. We demonstrate our method on a compilation of 3D brain MRI datasets, and show that it can learn high-quality templates that are representative of populations. We find that annotated conditional templates enable better registration than their unlabeled unconditional counterparts, and outperform other templates construction methods.
Classifying Phonotrauma Severity from Vocal Fold Images with Soft Ordinal Regression
Nov 12, 2025



Abstract:Phonotrauma refers to vocal fold tissue damage resulting from exposure to forces during voicing. It occurs on a continuum from mild to severe, and treatment options can vary based on severity. Assessment of severity involves a clinician's expert judgment, which is costly and can vary widely in reliability. In this work, we present the first method for automatically classifying phonotrauma severity from vocal fold images. To account for the ordinal nature of the labels, we adopt a widely used ordinal regression framework. To account for label uncertainty, we propose a novel modification to ordinal regression loss functions that enables them to operate on soft labels reflecting annotator rating distributions. Our proposed soft ordinal regression method achieves predictive performance approaching that of clinical experts, while producing well-calibrated uncertainty estimates. By providing an automated tool for phonotrauma severity assessment, our work can enable large-scale studies of phonotrauma, ultimately leading to improved clinical understanding and patient care.
MultiMorph: On-demand Atlas Construction
Mar 31, 2025Abstract:We present MultiMorph, a fast and efficient method for constructing anatomical atlases on the fly. Atlases capture the canonical structure of a collection of images and are essential for quantifying anatomical variability across populations. However, current atlas construction methods often require days to weeks of computation, thereby discouraging rapid experimentation. As a result, many scientific studies rely on suboptimal, precomputed atlases from mismatched populations, negatively impacting downstream analyses. MultiMorph addresses these challenges with a feedforward model that rapidly produces high-quality, population-specific atlases in a single forward pass for any 3D brain dataset, without any fine-tuning or optimization. MultiMorph is based on a linear group-interaction layer that aggregates and shares features within the group of input images. Further, by leveraging auxiliary synthetic data, MultiMorph generalizes to new imaging modalities and population groups at test-time. Experimentally, MultiMorph outperforms state-of-the-art optimization-based and learning-based atlas construction methods in both small and large population settings, with a 100-fold reduction in time. This makes MultiMorph an accessible framework for biomedical researchers without machine learning expertise, enabling rapid, high-quality atlas generation for diverse studies.
Shape-aware Segmentation of the Placenta in BOLD Fetal MRI Time Series
Dec 08, 2023



Abstract:Blood oxygen level dependent (BOLD) MRI time series with maternal hyperoxia can assess placental oxygenation and function. Measuring precise BOLD changes in the placenta requires accurate temporal placental segmentation and is confounded by fetal and maternal motion, contractions, and hyperoxia-induced intensity changes. Current BOLD placenta segmentation methods warp a manually annotated subject-specific template to the entire time series. However, as the placenta is a thin, elongated, and highly non-rigid organ subject to large deformations and obfuscated edges, existing work cannot accurately segment the placental shape, especially near boundaries. In this work, we propose a machine learning segmentation framework for placental BOLD MRI and apply it to segmenting each volume in a time series. We use a placental-boundary weighted loss formulation and perform a comprehensive evaluation across several popular segmentation objectives. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. Biomedically, our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. We further find that boundary-weighting increases placental segmentation performance by 8.3% and 6.0% Dice coefficient for the cross-entropy and signed distance transform objectives, respectively. Our code and trained model is available at https://github.com/mabulnaga/automatic-placenta-segmentation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:017. arXiv admin note: substantial text overlap with arXiv:2208.02895
Dynamic Neural Fields for Learning Atlases of 4D Fetal MRI Time-series
Nov 06, 2023


Abstract:We present a method for fast biomedical image atlas construction using neural fields. Atlases are key to biomedical image analysis tasks, yet conventional and deep network estimation methods remain time-intensive. In this preliminary work, we frame subject-specific atlas building as learning a neural field of deformable spatiotemporal observations. We apply our method to learning subject-specific atlases and motion stabilization of dynamic BOLD MRI time-series of fetuses in utero. Our method yields high-quality atlases of fetal BOLD time-series with $\sim$5-7$\times$ faster convergence compared to existing work. While our method slightly underperforms well-tuned baselines in terms of anatomical overlap, it estimates templates significantly faster, thus enabling rapid processing and stabilization of large databases of 4D dynamic MRI acquisitions. Code is available at https://github.com/Kidrauh/neural-atlasing
Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series
Oct 16, 2023Abstract:The placenta plays a crucial role in fetal development. Automated 3D placenta segmentation from fetal EPI MRI holds promise for advancing prenatal care. This paper proposes an effective semi-supervised learning method for improving placenta segmentation in fetal EPI MRI time series. We employ consistency regularization loss that promotes consistency under spatial transformation of the same image and temporal consistency across nearby images in a time series. The experimental results show that the method improves the overall segmentation accuracy and provides better performance for outliers and hard samples. The evaluation also indicates that our method improves the temporal coherency of the prediction, which could lead to more accurate computation of temporal placental biomarkers. This work contributes to the study of the placenta and prenatal clinical decision-making. Code is available at https://github.com/firstmover/cr-seg.
AnyStar: Domain randomized universal star-convex 3D instance segmentation
Jul 13, 2023Abstract:Star-convex shapes arise across bio-microscopy and radiology in the form of nuclei, nodules, metastases, and other units. Existing instance segmentation networks for such structures train on densely labeled instances for each dataset, which requires substantial and often impractical manual annotation effort. Further, significant reengineering or finetuning is needed when presented with new datasets and imaging modalities due to changes in contrast, shape, orientation, resolution, and density. We present AnyStar, a domain-randomized generative model that simulates synthetic training data of blob-like objects with randomized appearance, environments, and imaging physics to train general-purpose star-convex instance segmentation networks. As a result, networks trained using our generative model do not require annotated images from unseen datasets. A single network trained on our synthesized data accurately 3D segments C. elegans and P. dumerilii nuclei in fluorescence microscopy, mouse cortical nuclei in micro-CT, zebrafish brain nuclei in EM, and placental cotyledons in human fetal MRI, all without any retraining, finetuning, transfer learning, or domain adaptation. Code is available at https://github.com/neel-dey/AnyStar.
Automatic Segmentation of the Placenta in BOLD MRI Time Series
Aug 04, 2022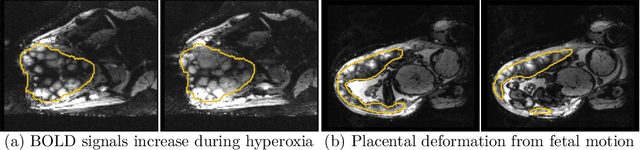
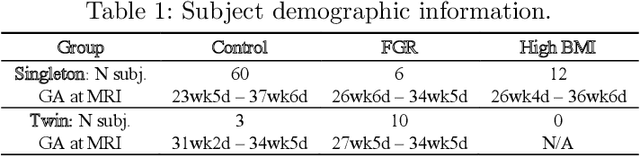
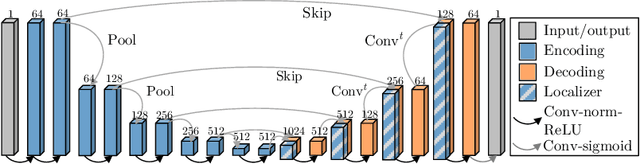

Abstract:Blood oxygen level dependent (BOLD) MRI with maternal hyperoxia can assess oxygen transport within the placenta and has emerged as a promising tool to study placental function. Measuring signal changes over time requires segmenting the placenta in each volume of the time series. Due to the large number of volumes in the BOLD time series, existing studies rely on registration to map all volumes to a manually segmented template. As the placenta can undergo large deformation due to fetal motion, maternal motion, and contractions, this approach often results in a large number of discarded volumes, where the registration approach fails. In this work, we propose a machine learning model based on a U-Net neural network architecture to automatically segment the placenta in BOLD MRI and apply it to segmenting each volume in a time series. We use a boundary-weighted loss function to accurately capture the placental shape. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. We achieve a Dice score of 0.83+/-0.04 when matching with ground truth labels and our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. Our code and trained model are available at https://github.com/mabulnaga/automatic-placenta-segmentation.
Symmetric Volume Maps
Feb 05, 2022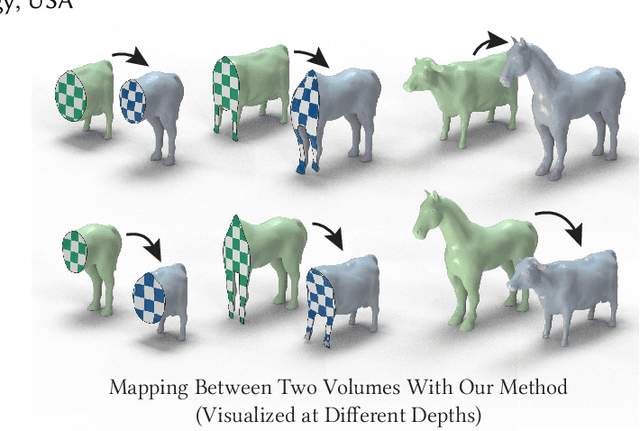
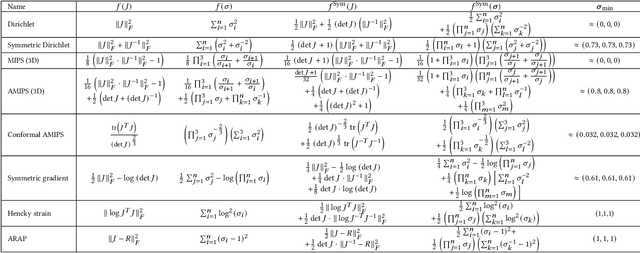
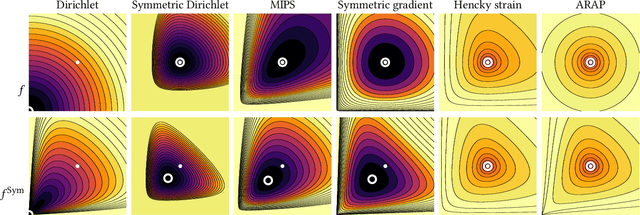
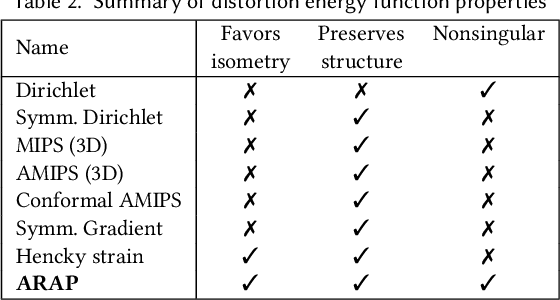
Abstract:Although shape correspondence is a central problem in geometry processing, most methods for this task apply only to two-dimensional surfaces. The neglected task of volumetric correspondence--a natural extension relevant to shapes extracted from simulation, medical imaging, volume rendering, and even improving surface maps of boundary representations--presents unique challenges that do not appear in the two-dimensional case. In this work, we propose a method for mapping between volumes represented as tetrahedral meshes. Our formulation minimizes a distortion energy designed to extract maps symmetrically, i.e., without dependence on the ordering of the source and target domains. We accompany our method with theoretical discussion describing the consequences of this symmetry assumption, leading us to select a symmetrized ARAP energy that favors isometric correspondences. Our final formulation optimizes for near-isometry while matching the boundary. We demonstrate our method on a diverse geometric dataset, producing low-distortion matchings that align to the boundary.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge