Dattesh Shanbhag
Automated Motion Artifact Check for MRI (AutoMAC-MRI): An Interpretable Framework for Motion Artifact Detection and Severity Assessment
Dec 17, 2025Abstract:Motion artifacts degrade MRI image quality and increase patient recalls. Existing automated quality assessment methods are largely limited to binary decisions and provide little interpretability. We introduce AutoMAC-MRI, an explainable framework for grading motion artifacts across heterogeneous MR contrasts and orientations. The approach uses supervised contrastive learning to learn a discriminative representation of motion severity. Within this feature space, we compute grade-specific affinity scores that quantify an image's proximity to each motion grade, thereby making grade assignments transparent and interpretable. We evaluate AutoMAC-MRI on more than 5000 expert-annotated brain MRI slices spanning multiple contrasts and views. Experiments assessing affinity scores against expert labels show that the scores align well with expert judgment, supporting their use as an interpretable measure of motion severity. By coupling accurate grade detection with per-grade affinity scoring, AutoMAC-MRI enables inline MRI quality control, with the potential to reduce unnecessary rescans and improve workflow efficiency.
Test Time Optimized Generalized AI-based Medical Image Registration Method
Dec 16, 2025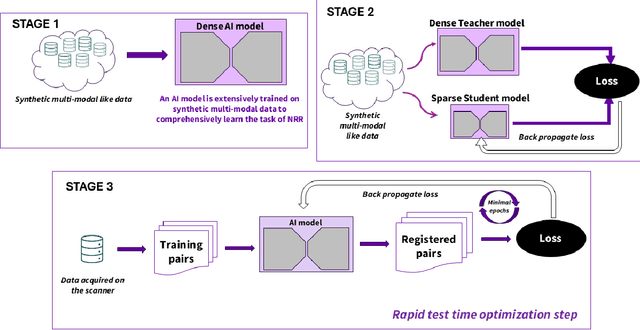
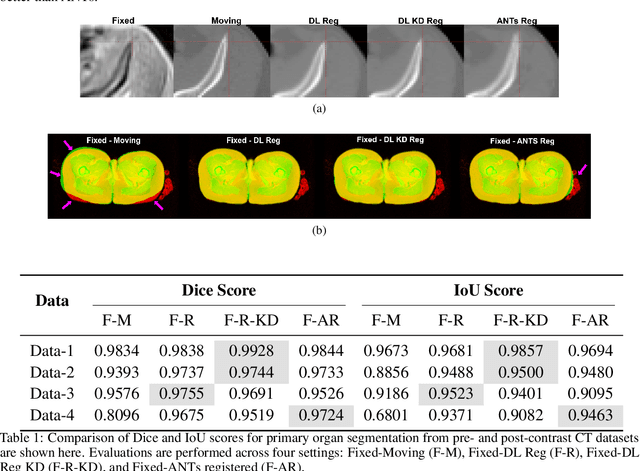
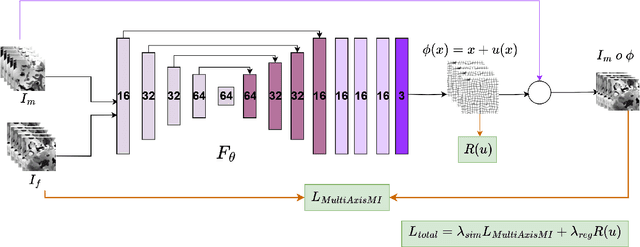
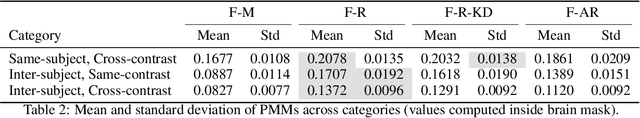
Abstract:Medical image registration is critical for aligning anatomical structures across imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound. Among existing techniques, non-rigid registration (NRR) is particularly challenging due to the need to capture complex anatomical deformations caused by physiological processes like respiration or contrast-induced signal variations. Traditional NRR methods, while theoretically robust, often require extensive parameter tuning and incur high computational costs, limiting their use in real-time clinical workflows. Recent deep learning (DL)-based approaches have shown promise; however, their dependence on task-specific retraining restricts scalability and adaptability in practice. These limitations underscore the need for efficient, generalizable registration frameworks capable of handling heterogeneous imaging contexts. In this work, we introduce a novel AI-driven framework for 3D non-rigid registration that generalizes across multiple imaging modalities and anatomical regions. Unlike conventional methods that rely on application-specific models, our approach eliminates anatomy- or modality-specific customization, enabling streamlined integration into diverse clinical environments.
Data Adaptive Few-shot Multi Label Segmentation with Foundation Model
Oct 13, 2024



Abstract:The high cost of obtaining accurate annotations for image segmentation and localization makes the use of one and few shot algorithms attractive. Several state-of-the-art methods for few-shot segmentation have emerged, including text-based prompting for the task but suffer from sub-optimal performance for medical images. Leveraging sub-pixel level features of existing Vision Transformer (ViT) based foundation models for identifying similar region of interest (RoI) based on a single template image have been shown to be very effective for one shot segmentation and localization in medical images across modalities. However, such methods rely on assumption that template image and test image are well matched and simple correlation is sufficient to obtain correspondences. In practice, however such an approach can fail to generalize in clinical data due to patient pose changes, inter-protocol variations even within a single modality or extend to 3D data using single template image. Moreover, for multi-label tasks, the RoI identification has to be performed sequentially. In this work, we propose foundation model (FM) based adapters for single label, multi-label localization and segmentation to address these concerns. We demonstrate the efficacy of the proposed method for multiple segmentation and localization tasks for both 2D and 3D data as we well as clinical data with different poses and evaluate against the state of the art few shot segmentation methods.
Language Augmentation in CLIP for Improved Anatomy Detection on Multi-modal Medical Images
May 31, 2024



Abstract:Vision-language models have emerged as a powerful tool for previously challenging multi-modal classification problem in the medical domain. This development has led to the exploration of automated image description generation for multi-modal clinical scans, particularly for radiology report generation. Existing research has focused on clinical descriptions for specific modalities or body regions, leaving a gap for a model providing entire-body multi-modal descriptions. In this paper, we address this gap by automating the generation of standardized body station(s) and list of organ(s) across the whole body in multi-modal MR and CT radiological images. Leveraging the versatility of the Contrastive Language-Image Pre-training (CLIP), we refine and augment the existing approach through multiple experiments, including baseline model fine-tuning, adding station(s) as a superset for better correlation between organs, along with image and language augmentations. Our proposed approach demonstrates 47.6% performance improvement over baseline PubMedCLIP.
Region of Interest focused MRI to Synthetic CT Translation using Regression and Classification Multi-task Network
Mar 30, 2022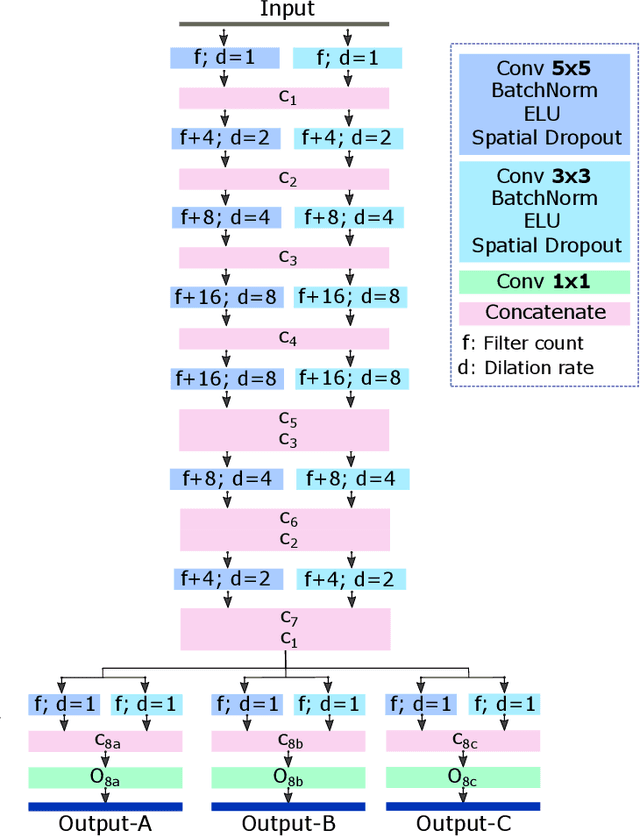
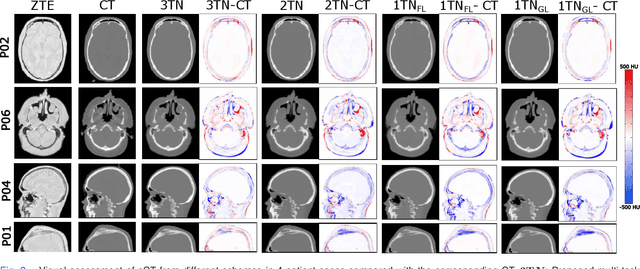
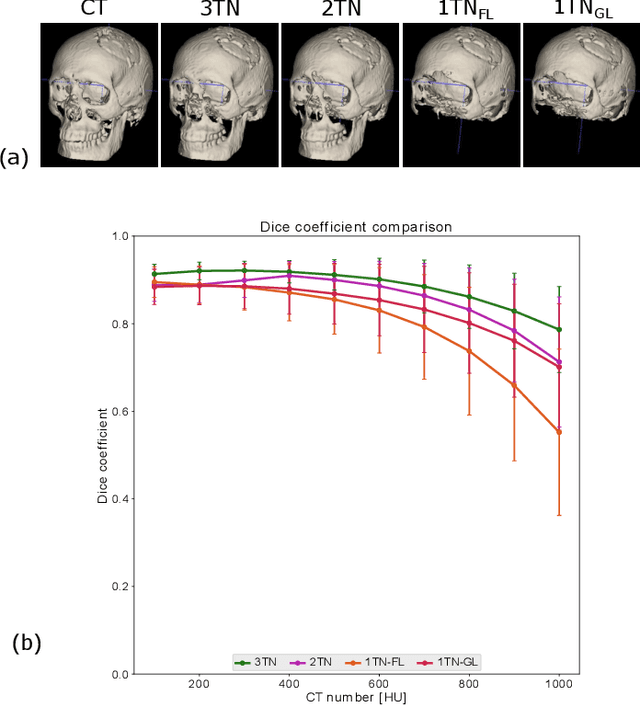
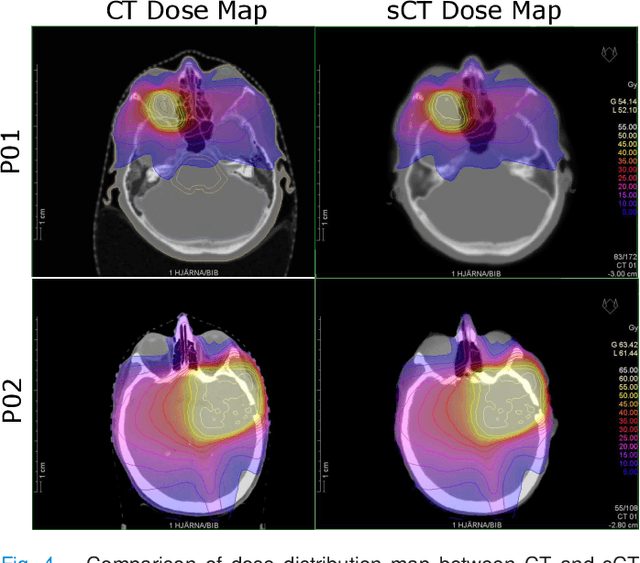
Abstract:In this work, we present a method for synthetic CT (sCT) generation from zero-echo-time (ZTE) MRI aimed at structural and quantitative accuracies of the image, with a particular focus on the accurate bone density value prediction. We propose a loss function that favors a spatially sparse region in the image. We harness the ability of a multi-task network to produce correlated outputs as a framework to enable localisation of region of interest (RoI) via classification, emphasize regression of values within RoI and still retain the overall accuracy via global regression. The network is optimized by a composite loss function that combines a dedicated loss from each task. We demonstrate how the multi-task network with RoI focused loss offers an advantage over other configurations of the network to achieve higher accuracy of performance. This is relevant to sCT where failure to accurately estimate high Hounsfield Unit values of bone could lead to impaired accuracy in clinical applications. We compare the dose calculation maps from the proposed sCT and the real CT in a radiation therapy treatment planning setup.
An SVM Based Approach for Cardiac View Planning
Jul 11, 2014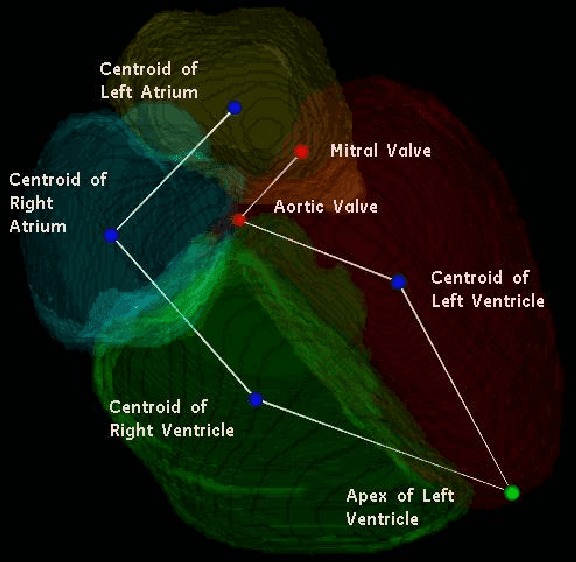
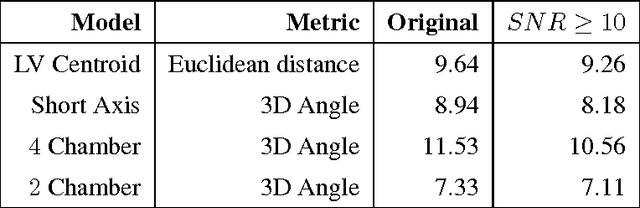
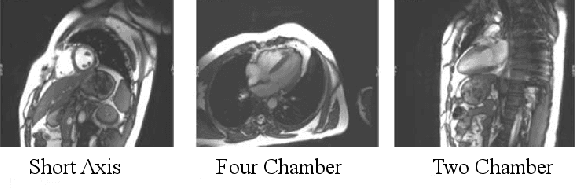
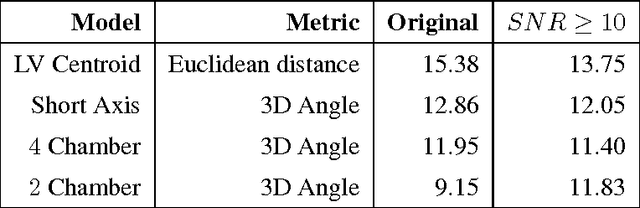
Abstract:We consider the problem of automatically prescribing oblique planes (short axis, 4 chamber and 2 chamber views) in Cardiac Magnetic Resonance Imaging (MRI). A concern with technologist-driven acquisitions of these planes is the quality and time taken for the total examination. We propose an automated solution incorporating anatomical features external to the cardiac region. The solution uses support vector machine regression models wherein complexity and feature selection are optimized using multi-objective genetic algorithms. Additionally, we examine the robustness of our approach by training our models on images with additive Rician-Gaussian mixtures at varying Signal to Noise (SNR) levels. Our approach has shown promising results, with an angular deviation of less than 15 degrees on 90% cases across oblique planes, measured in terms of average 6-fold cross validation performance -- this is generally within acceptable bounds of variation as specified by clinicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge