Zhoubing Xu
IntraStyler: Exemplar-based Style Synthesis for Cross-modality Domain Adaptation
Jan 01, 2026Abstract:Image-level domain alignment is the de facto approach for unsupervised domain adaptation, where unpaired image translation is used to minimize the domain gap. Prior studies mainly focus on the domain shift between the source and target domains, whereas the intra-domain variability remains under-explored. To address the latter, an effective strategy is to diversify the styles of the synthetic target domain data during image translation. However, previous methods typically require intra-domain variations to be pre-specified for style synthesis, which may be impractical. In this paper, we propose an exemplar-based style synthesis method named IntraStyler, which can capture diverse intra-domain styles without any prior knowledge. Specifically, IntraStyler uses an exemplar image to guide the style synthesis such that the output style matches the exemplar style. To extract the style-only features, we introduce a style encoder to learn styles discriminatively based on contrastive learning. We evaluate the proposed method on the largest public dataset for cross-modality domain adaptation, CrossMoDA 2023. Our experiments show the efficacy of our method in controllable style synthesis and the benefits of diverse synthetic data for downstream segmentation. Code is available at https://github.com/han-liu/IntraStyler.
Predicting fluorescent labels in label-free microscopy images with pix2pix and adaptive loss in Light My Cells challenge
Jun 22, 2024



Abstract:Fluorescence labeling is the standard approach to reveal cellular structures and other subcellular constituents for microscopy images. However, this invasive procedure may perturb or even kill the cells and the procedure itself is highly time-consuming and complex. Recently, in silico labeling has emerged as a promising alternative, aiming to use machine learning models to directly predict the fluorescently labeled images from label-free microscopy. In this paper, we propose a deep learning-based in silico labeling method for the Light My Cells challenge. Built upon pix2pix, our proposed method can be trained using the partially labeled datasets with an adaptive loss. Moreover, we explore the effectiveness of several training strategies to handle different input modalities, such as training them together or separately. The results show that our method achieves promising performance for in silico labeling. Our code is available at https://github.com/MedICL-VU/LightMyCells.
Learning Site-specific Styles for Multi-institutional Unsupervised Cross-modality Domain Adaptation
Nov 22, 2023



Abstract:Unsupervised cross-modality domain adaptation is a challenging task in medical image analysis, and it becomes more challenging when source and target domain data are collected from multiple institutions. In this paper, we present our solution to tackle the multi-institutional unsupervised domain adaptation for the crossMoDA 2023 challenge. First, we perform unpaired image translation to translate the source domain images to the target domain, where we design a dynamic network to generate synthetic target domain images with controllable, site-specific styles. Afterwards, we train a segmentation model using the synthetic images and further reduce the domain gap by self-training. Our solution achieved the 1st place during both the validation and testing phases of the challenge. The code repository is publicly available at https://github.com/MedICL-VU/crossmoda2023.
COLosSAL: A Benchmark for Cold-start Active Learning for 3D Medical Image Segmentation
Jul 22, 2023Abstract:Medical image segmentation is a critical task in medical image analysis. In recent years, deep learning based approaches have shown exceptional performance when trained on a fully-annotated dataset. However, data annotation is often a significant bottleneck, especially for 3D medical images. Active learning (AL) is a promising solution for efficient annotation but requires an initial set of labeled samples to start active selection. When the entire data pool is unlabeled, how do we select the samples to annotate as our initial set? This is also known as the cold-start AL, which permits only one chance to request annotations from experts without access to previously annotated data. Cold-start AL is highly relevant in many practical scenarios but has been under-explored, especially for 3D medical segmentation tasks requiring substantial annotation effort. In this paper, we present a benchmark named COLosSAL by evaluating six cold-start AL strategies on five 3D medical image segmentation tasks from the public Medical Segmentation Decathlon collection. We perform a thorough performance analysis and explore important open questions for cold-start AL, such as the impact of budget on different strategies. Our results show that cold-start AL is still an unsolved problem for 3D segmentation tasks but some important trends have been observed. The code repository, data partitions, and baseline results for the complete benchmark are publicly available at https://github.com/MedICL-VU/COLosSAL.
COSST: Multi-organ Segmentation with Partially Labeled Datasets Using Comprehensive Supervisions and Self-training
Apr 28, 2023Abstract:Deep learning models have demonstrated remarkable success in multi-organ segmentation but typically require large-scale datasets with all organs of interest annotated. However, medical image datasets are often low in sample size and only partially labeled, i.e., only a subset of organs are annotated. Therefore, it is crucial to investigate how to learn a unified model on the available partially labeled datasets to leverage their synergistic potential. In this paper, we empirically and systematically study the partial-label segmentation with in-depth analyses on the existing approaches and identify three distinct types of supervision signals, including two signals derived from ground truth and one from pseudo label. We propose a novel training framework termed COSST, which effectively and efficiently integrates comprehensive supervision signals with self-training. Concretely, we first train an initial unified model using two ground truth-based signals and then iteratively incorporate the pseudo label signal to the initial model using self-training. To mitigate performance degradation caused by unreliable pseudo labels, we assess the reliability of pseudo labels via outlier detection in latent space and exclude the most unreliable pseudo labels from each self-training iteration. Extensive experiments are conducted on six CT datasets for three partial-label segmentation tasks. Experimental results show that our proposed COSST achieves significant improvement over the baseline method, i.e., individual networks trained on each partially labeled dataset. Compared to the state-of-the-art partial-label segmentation methods, COSST demonstrates consistent superior performance on various segmentation tasks and with different training data size.
UNesT: Local Spatial Representation Learning with Hierarchical Transformer for Efficient Medical Segmentation
Sep 28, 2022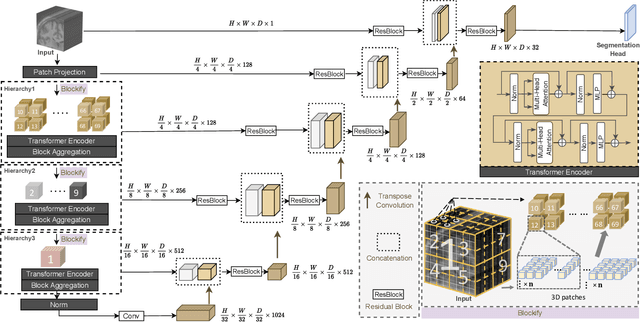
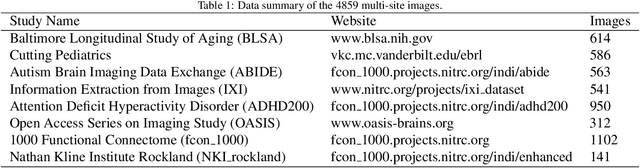
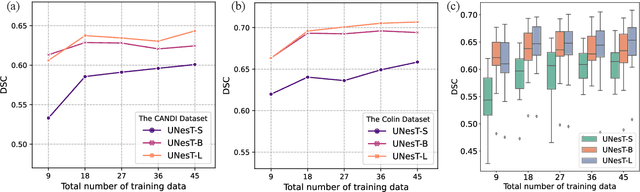
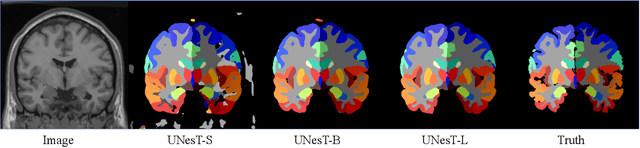
Abstract:Transformer-based models, capable of learning better global dependencies, have recently demonstrated exceptional representation learning capabilities in computer vision and medical image analysis. Transformer reformats the image into separate patches and realize global communication via the self-attention mechanism. However, positional information between patches is hard to preserve in such 1D sequences, and loss of it can lead to sub-optimal performance when dealing with large amounts of heterogeneous tissues of various sizes in 3D medical image segmentation. Additionally, current methods are not robust and efficient for heavy-duty medical segmentation tasks such as predicting a large number of tissue classes or modeling globally inter-connected tissues structures. Inspired by the nested hierarchical structures in vision transformer, we proposed a novel 3D medical image segmentation method (UNesT), employing a simplified and faster-converging transformer encoder design that achieves local communication among spatially adjacent patch sequences by aggregating them hierarchically. We extensively validate our method on multiple challenging datasets, consisting anatomies of 133 structures in brain, 14 organs in abdomen, 4 hierarchical components in kidney, and inter-connected kidney tumors). We show that UNesT consistently achieves state-of-the-art performance and evaluate its generalizability and data efficiency. Particularly, the model achieves whole brain segmentation task complete ROI with 133 tissue classes in single network, outperforms prior state-of-the-art method SLANT27 ensembled with 27 network tiles, our model performance increases the mean DSC score of the publicly available Colin and CANDI dataset from 0.7264 to 0.7444 and from 0.6968 to 0.7025, respectively.
A Comparative Study of Confidence Calibration in Deep Learning: From Computer Vision to Medical Imaging
Jun 17, 2022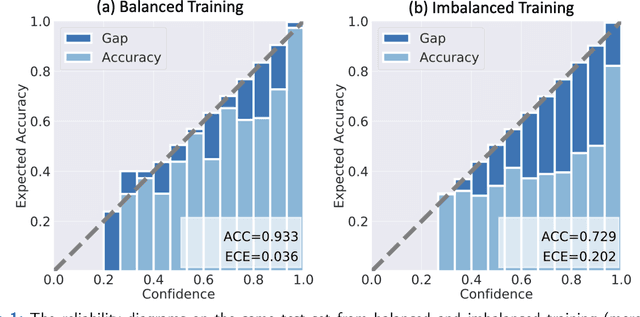
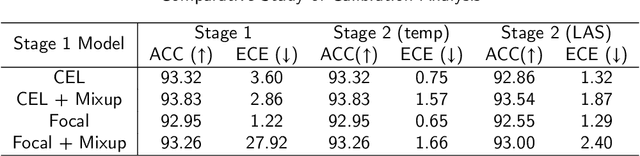
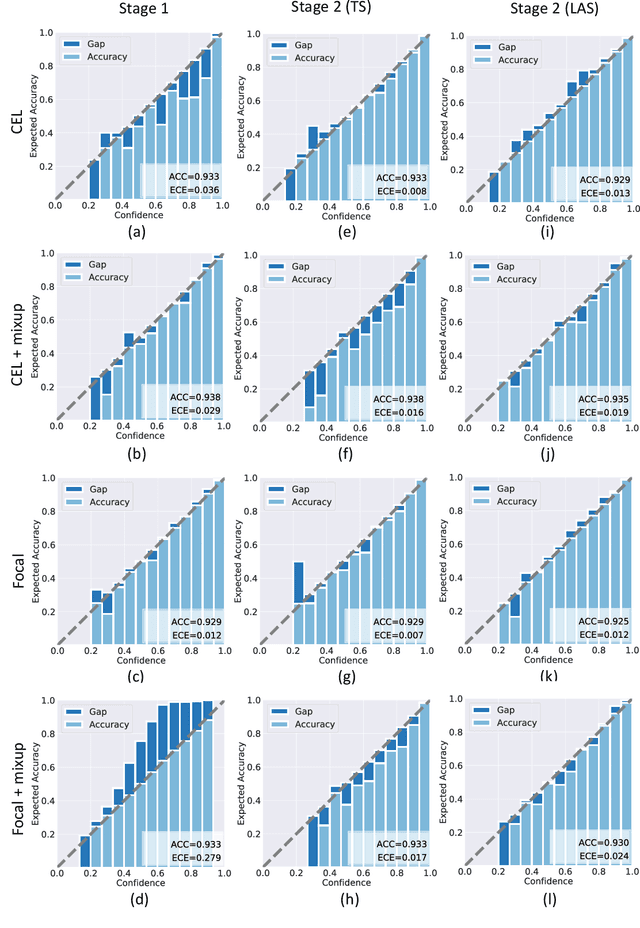
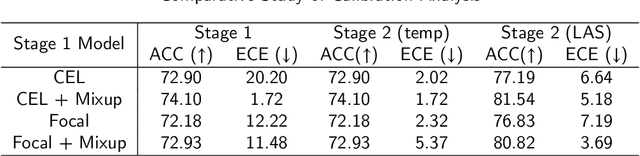
Abstract:Although deep learning prediction models have been successful in the discrimination of different classes, they can often suffer from poor calibration across challenging domains including healthcare. Moreover, the long-tail distribution poses great challenges in deep learning classification problems including clinical disease prediction. There are approaches proposed recently to calibrate deep prediction in computer vision, but there are no studies found to demonstrate how the representative models work in different challenging contexts. In this paper, we bridge the confidence calibration from computer vision to medical imaging with a comparative study of four high-impact calibration models. Our studies are conducted in different contexts (natural image classification and lung cancer risk estimation) including in balanced vs. imbalanced training sets and in computer vision vs. medical imaging. Our results support key findings: (1) We achieve new conclusions which are not studied under different learning contexts, e.g., combining two calibration models that both mitigate the overconfident prediction can lead to under-confident prediction, and simpler calibration models from the computer vision domain tend to be more generalizable to medical imaging. (2) We highlight the gap between general computer vision tasks and medical imaging prediction, e.g., calibration methods ideal for general computer vision tasks may in fact damage the calibration of medical imaging prediction. (3) We also reinforce previous conclusions in natural image classification settings. We believe that this study has merits to guide readers to choose calibration models and understand gaps between general computer vision and medical imaging domains.
Characterizing Renal Structures with 3D Block Aggregate Transformers
Mar 04, 2022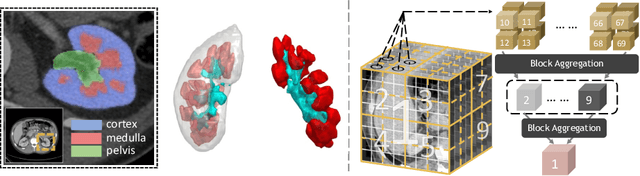
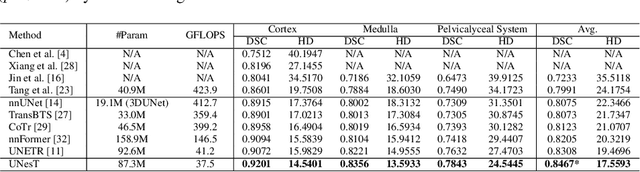
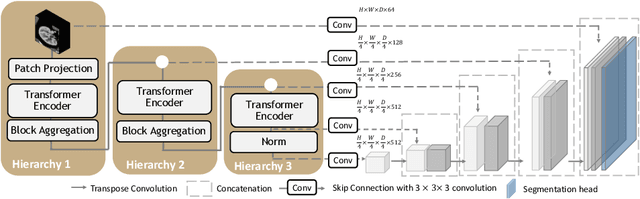
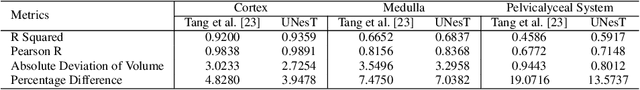
Abstract:Efficiently quantifying renal structures can provide distinct spatial context and facilitate biomarker discovery for kidney morphology. However, the development and evaluation of the transformer model to segment the renal cortex, medulla, and collecting system remains challenging due to data inefficiency. Inspired by the hierarchical structures in vision transformer, we propose a novel method using a 3D block aggregation transformer for segmenting kidney components on contrast-enhanced CT scans. We construct the first cohort of renal substructures segmentation dataset with 116 subjects under institutional review board (IRB) approval. Our method yields the state-of-the-art performance (Dice of 0.8467) against the baseline approach of 0.8308 with the data-efficient design. The Pearson R achieves 0.9891 between the proposed method and manual standards and indicates the strong correlation and reproducibility for volumetric analysis. We extend the proposed method to the public KiTS dataset, the method leads to improved accuracy compared to transformer-based approaches. We show that the 3D block aggregation transformer can achieve local communication between sequence representations without modifying self-attention, and it can serve as an accurate and efficient quantification tool for characterizing renal structures.
3D Tomographic Pattern Synthesis for Enhancing the Quantification of COVID-19
May 05, 2020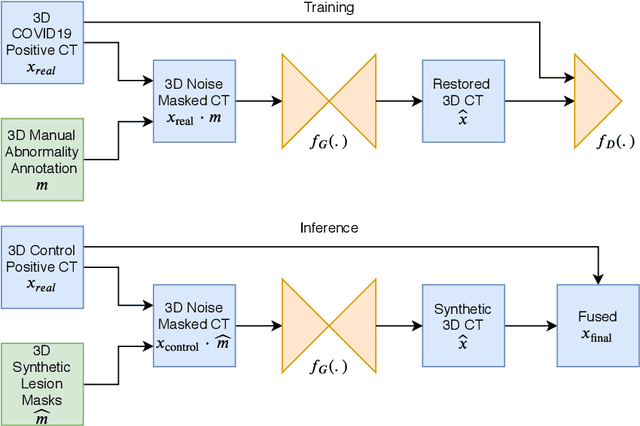
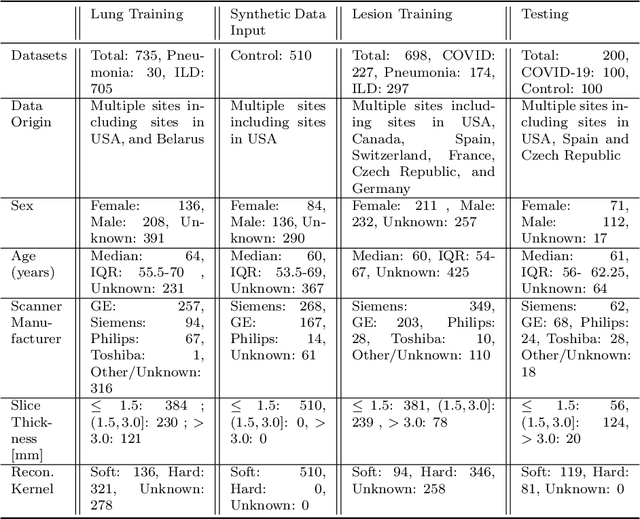
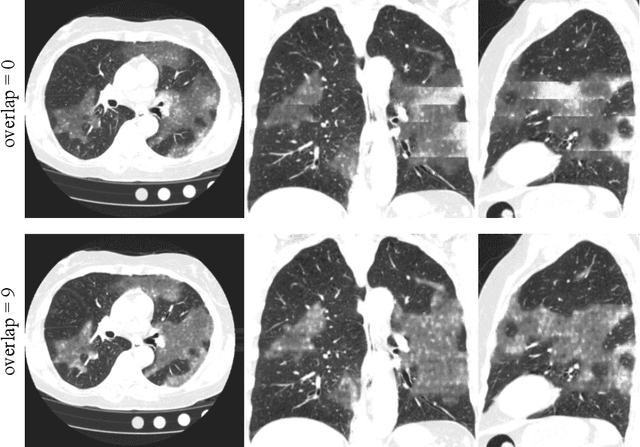
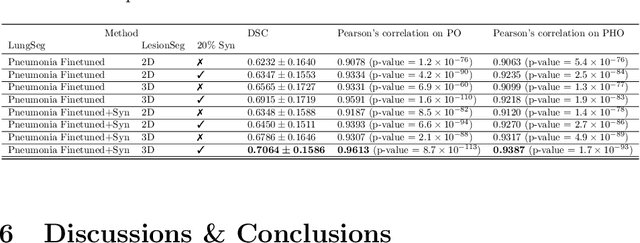
Abstract:The Coronavirus Disease (COVID-19) has affected 1.8 million people and resulted in more than 110,000 deaths as of April 12, 2020. Several studies have shown that tomographic patterns seen on chest Computed Tomography (CT), such as ground-glass opacities, consolidations, and crazy paving pattern, are correlated with the disease severity and progression. CT imaging can thus emerge as an important modality for the management of COVID-19 patients. AI-based solutions can be used to support CT based quantitative reporting and make reading efficient and reproducible if quantitative biomarkers, such as the Percentage of Opacity (PO), can be automatically computed. However, COVID-19 has posed unique challenges to the development of AI, specifically concerning the availability of appropriate image data and annotations at scale. In this paper, we propose to use synthetic datasets to augment an existing COVID-19 database to tackle these challenges. We train a Generative Adversarial Network (GAN) to inpaint COVID-19 related tomographic patterns on chest CTs from patients without infectious diseases. Additionally, we leverage location priors derived from manually labeled COVID-19 chest CTs patients to generate appropriate abnormality distributions. Synthetic data are used to improve both lung segmentation and segmentation of COVID-19 patterns by adding 20% of synthetic data to the real COVID-19 training data. We collected 2143 chest CTs, containing 327 COVID-19 positive cases, acquired from 12 sites across 7 countries. By testing on 100 COVID-19 positive and 100 control cases, we show that synthetic data can help improve both lung segmentation (+6.02% lesion inclusion rate) and abnormality segmentation (+2.78% dice coefficient), leading to an overall more accurate PO computation (+2.82% Pearson coefficient).
Quantification of Tomographic Patterns associated with COVID-19 from Chest CT
Apr 28, 2020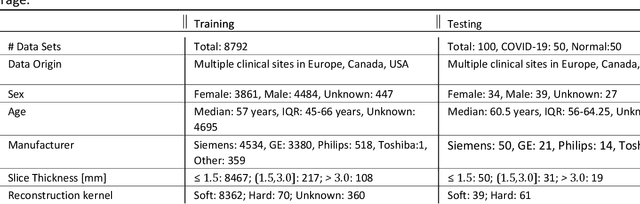
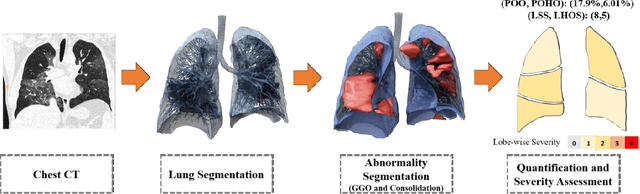
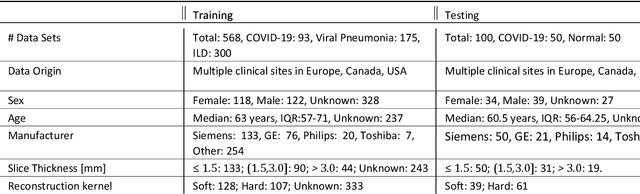
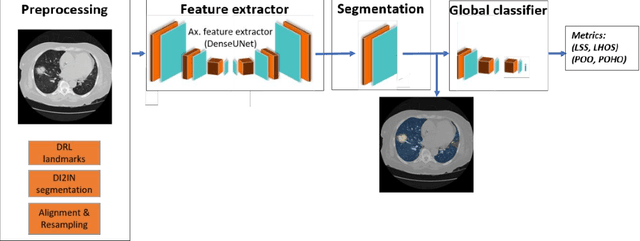
Abstract:Purpose: To present a method that automatically detects and quantifies abnormal tomographic patterns commonly present in COVID-19, namely Ground Glass Opacities (GGO) and consolidations. Given that high opacity abnormalities (i.e., consolidations) were shown to correlate with severe disease, the paper introduces two combined severity measures (Percentage of Opacity, Percentage of High Opacity) and (Lung Severity Score, Lung High Opacity Score). They quantify the extent of overall COVID-19 abnormalities and the presence of high opacity abnormalities, global and lobe-wise, respectively, being computed based on 3D segmentations of lesions, lungs, and lobes. Materials and Methods: The proposed method takes as input a non-contrasted Chest CT and segments the lesions, lungs, and lobes in 3D. It outputs two combined measures of the severity of lung/lobe involvement, quantifying both the extent of COVID-19 abnormalities and presence of high opacities, based on deep learning and deep reinforcement learning. The first measure (POO, POHO) is global, while the second (LSS, LHOS) is lobe-wise. Evaluation is reported on CTs of 100 subjects (50 COVID-19 confirmed and 50 controls) from institutions from Canada, Europe and US. Ground truth is established by manual annotations of lesions, lungs, and lobes. Results: Pearson Correlation Coefficient between method prediction and ground truth is 0.97 (POO), 0.98 (POHO), 0.96 (LSS), 0.96 (LHOS). Automated processing time to compute the severity scores is 10 seconds/case vs 30 mins needed for manual annotations. Conclusion: A new method identifies regions of abnormalities seen in COVID-19 non-contrasted Chest CT and computes (POO, POHO) and (LSS, LHOS) severity scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge