Learning Geometrically Disentangled Representations of Protein Folding Simulations
May 20, 2022N. Joseph Tatro, Payel Das, Pin-Yu Chen, Vijil Chenthamarakshan, Rongjie Lai
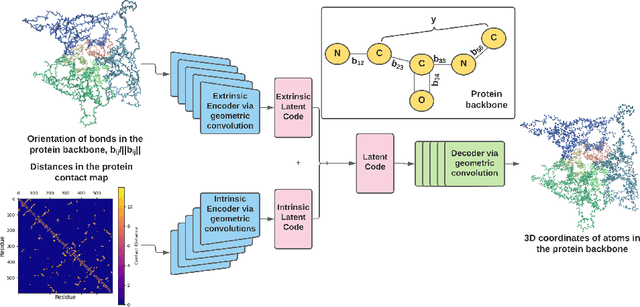
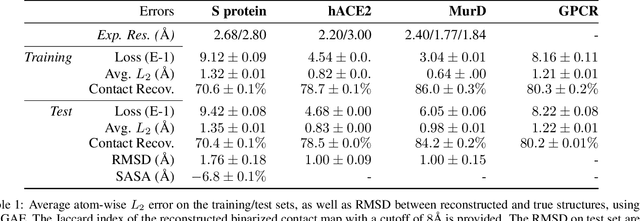
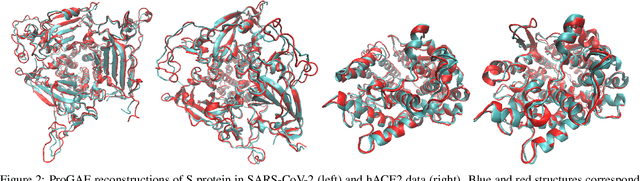

Massive molecular simulations of drug-target proteins have been used as a tool to understand disease mechanism and develop therapeutics. This work focuses on learning a generative neural network on a structural ensemble of a drug-target protein, e.g. SARS-CoV-2 Spike protein, obtained from computationally expensive molecular simulations. Model tasks involve characterizing the distinct structural fluctuations of the protein bound to various drug molecules, as well as efficient generation of protein conformations that can serve as an complement of a molecular simulation engine. Specifically, we present a geometric autoencoder framework to learn separate latent space encodings of the intrinsic and extrinsic geometries of the protein structure. For this purpose, the proposed Protein Geometric AutoEncoder (ProGAE) model is trained on the protein contact map and the orientation of the backbone bonds of the protein. Using ProGAE latent embeddings, we reconstruct and generate the conformational ensemble of a protein at or near the experimental resolution, while gaining better interpretability and controllability in term of protein structure generation from the learned latent space. Additionally, ProGAE models are transferable to a different state of the same protein or to a new protein of different size, where only the dense layer decoding from the latent representation needs to be retrained. Results show that our geometric learning-based method enjoys both accuracy and efficiency for generating complex structural variations, charting the path toward scalable and improved approaches for analyzing and enhancing high-cost simulations of drug-target proteins.
Accelerating Inhibitor Discovery for Multiple SARS-CoV-2 Targets with a Single, Sequence-Guided Deep Generative Framework
Apr 19, 2022Vijil Chenthamarakshan, Samuel C. Hoffman, C. David Owen, Petra Lukacik, Claire Strain-Damerell, Daren Fearon, Tika R. Malla, Anthony Tumber, Christopher J. Schofield, Helen M. E. Duyvesteyn, Wanwisa Dejnirattisai, Loic Carrique, Thomas S. Walter, Gavin R. Screaton, Tetiana Matviiuk, Aleksandra Mojsilovic, Jason Crain, Martin A. Walsh, David I. Stuart, Payel Das
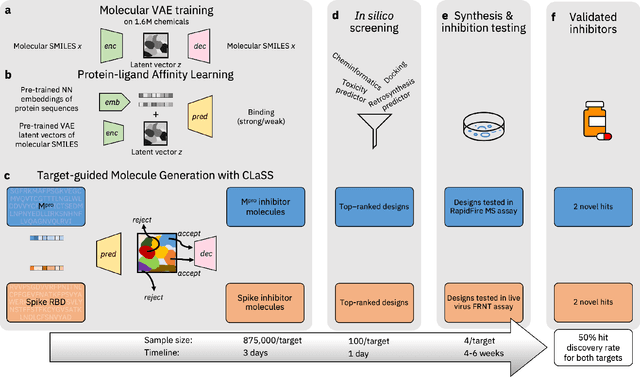


The COVID-19 pandemic has highlighted the urgency for developing more efficient molecular discovery pathways. As exhaustive exploration of the vast chemical space is infeasible, discovering novel inhibitor molecules for emerging drug-target proteins is challenging, particularly for targets with unknown structure or ligands. We demonstrate the broad utility of a single deep generative framework toward discovering novel drug-like inhibitor molecules against two distinct SARS-CoV-2 targets -- the main protease (Mpro) and the receptor binding domain (RBD) of the spike protein. To perform target-aware design, the framework employs a target sequence-conditioned sampling of novel molecules from a generative model. Micromolar-level in vitro inhibition was observed for two candidates (out of four synthesized) for each target. The most potent spike RBD inhibitor also emerged as a rare non-covalent antiviral with broad-spectrum activity against several SARS-CoV-2 variants in live virus neutralization assays. These results show a broadly deployable machine intelligence framework can accelerate hit discovery across different emerging drug-targets.
Protein Representation Learning by Geometric Structure Pretraining
Mar 14, 2022Zuobai Zhang, Minghao Xu, Arian Jamasb, Vijil Chenthamarakshan, Aurelie Lozano, Payel Das, Jian Tang
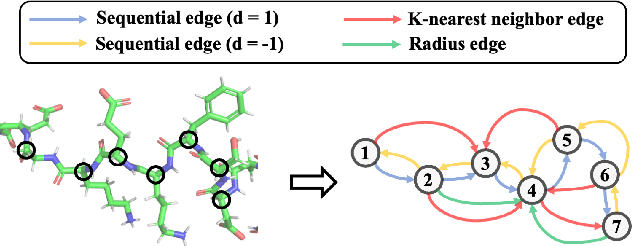
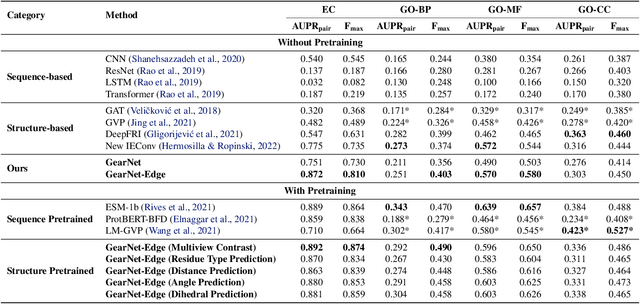
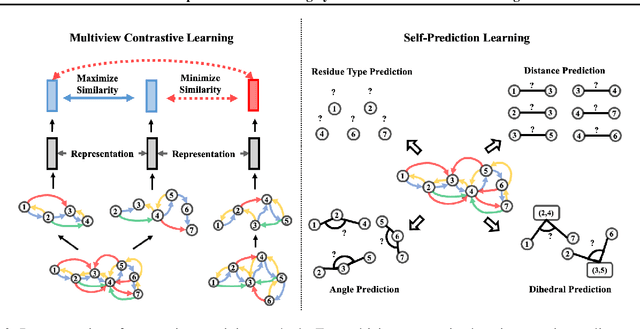
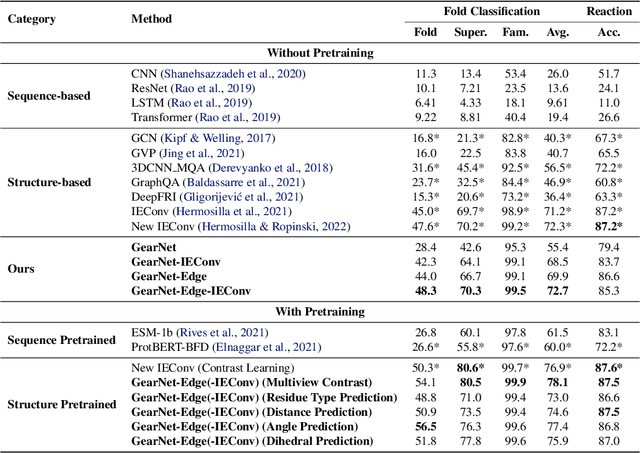
Learning effective protein representations is critical in a variety of tasks in biology such as predicting protein function or structure. Existing approaches usually pretrain protein language models on a large number of unlabeled amino acid sequences and then finetune the models with some labeled data in downstream tasks. Despite the effectiveness of sequence-based approaches, the power of pretraining on smaller numbers of known protein structures has not been explored for protein property prediction, though protein structures are known to be determinants of protein function. We first present a simple yet effective encoder to learn protein geometry features. We pretrain the protein graph encoder by leveraging multiview contrastive learning and different self-prediction tasks. Experimental results on both function prediction and fold classification tasks show that our proposed pretraining methods outperform or are on par with the state-of-the-art sequence-based methods using much less data. All codes and models will be published upon acceptance.
Sample-Efficient Generation of Novel Photo-acid Generator Molecules using a Deep Generative Model
Dec 02, 2021Samuel C. Hoffman, Vijil Chenthamarakshan, Dmitry Yu. Zubarev, Daniel P. Sanders, Payel Das
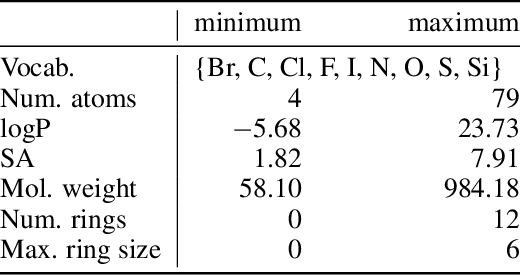
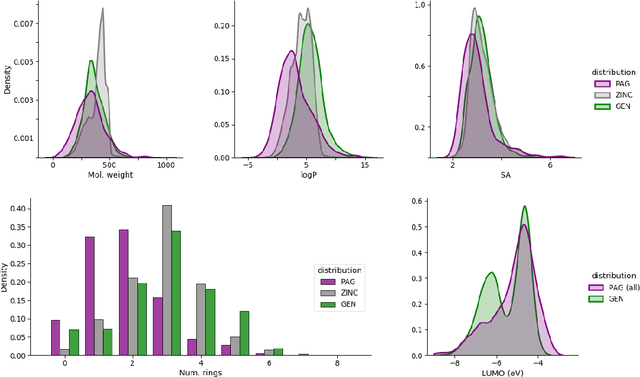
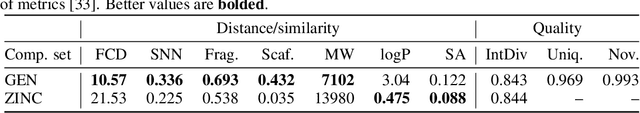
Photo-acid generators (PAGs) are compounds that release acids ($H^+$ ions) when exposed to light. These compounds are critical components of the photolithography processes that are used in the manufacture of semiconductor logic and memory chips. The exponential increase in the demand for semiconductors has highlighted the need for discovering novel photo-acid generators. While de novo molecule design using deep generative models has been widely employed for drug discovery and material design, its application to the creation of novel photo-acid generators poses several unique challenges, such as lack of property labels. In this paper, we highlight these challenges and propose a generative modeling approach that utilizes conditional generation from a pre-trained deep autoencoder and expert-in-the-loop techniques. The validity of the proposed approach was evaluated with the help of subject matter experts, indicating the promise of such an approach for applications beyond the creation of novel photo-acid generators.
Benchmarking deep generative models for diverse antibody sequence design
Nov 12, 2021Igor Melnyk, Payel Das, Vijil Chenthamarakshan, Aurelie Lozano
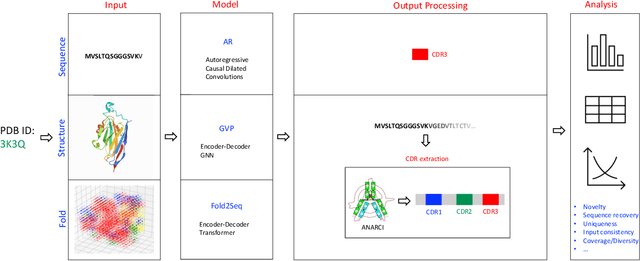

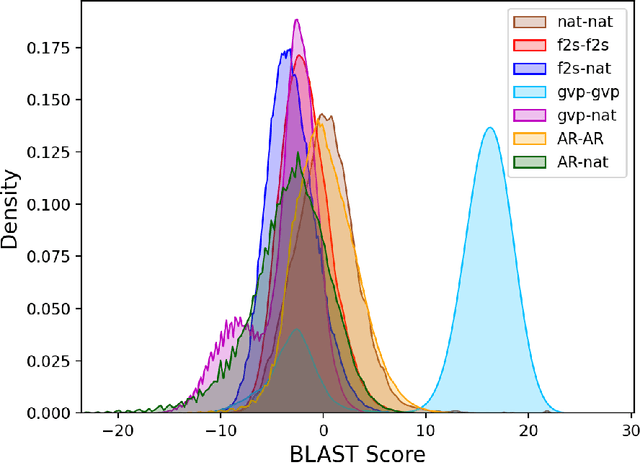
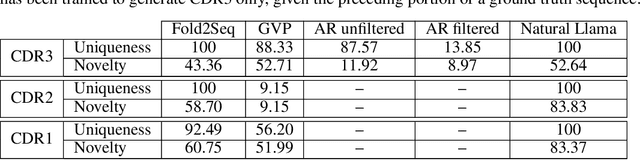
Computational protein design, i.e. inferring novel and diverse protein sequences consistent with a given structure, remains a major unsolved challenge. Recently, deep generative models that learn from sequences alone or from sequences and structures jointly have shown impressive performance on this task. However, those models appear limited in terms of modeling structural constraints, capturing enough sequence diversity, or both. Here we consider three recently proposed deep generative frameworks for protein design: (AR) the sequence-based autoregressive generative model, (GVP) the precise structure-based graph neural network, and Fold2Seq that leverages a fuzzy and scale-free representation of a three-dimensional fold, while enforcing structure-to-sequence (and vice versa) consistency. We benchmark these models on the task of computational design of antibody sequences, which demand designing sequences with high diversity for functional implication. The Fold2Seq framework outperforms the two other baselines in terms of diversity of the designed sequences, while maintaining the typical fold.
Fold2Seq: A Joint Sequence(1D)-Fold(3D) Embedding-based Generative Model for Protein Design
Jun 24, 2021Yue Cao, Payel Das, Vijil Chenthamarakshan, Pin-Yu Chen, Igor Melnyk, Yang Shen
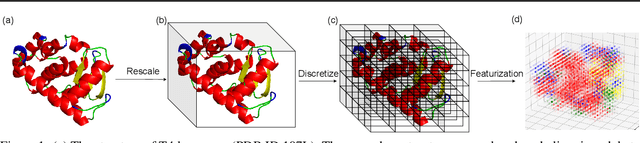
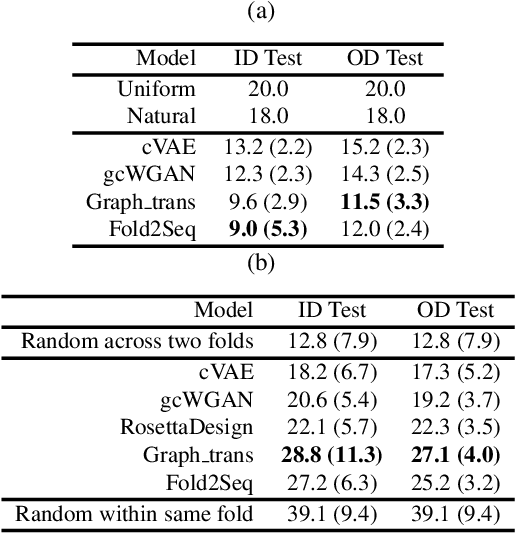
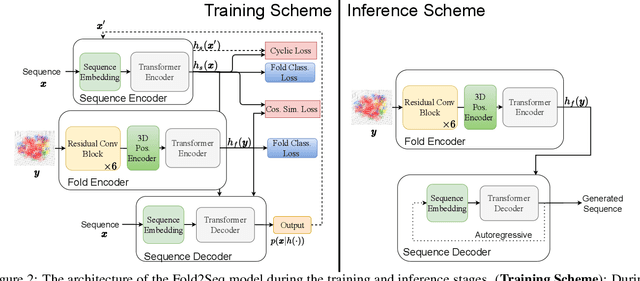
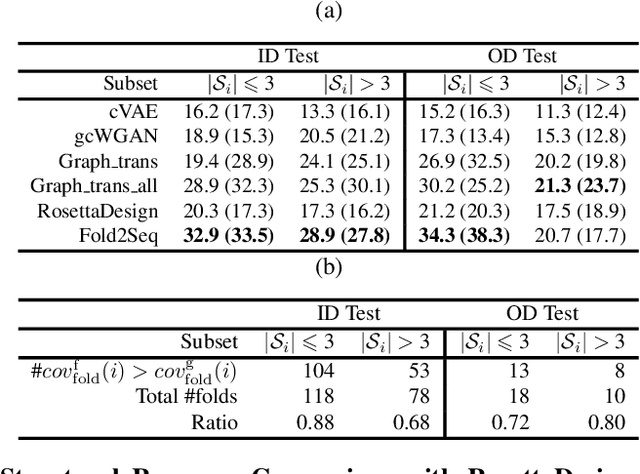
Designing novel protein sequences for a desired 3D topological fold is a fundamental yet non-trivial task in protein engineering. Challenges exist due to the complex sequence--fold relationship, as well as the difficulties to capture the diversity of the sequences (therefore structures and functions) within a fold. To overcome these challenges, we propose Fold2Seq, a novel transformer-based generative framework for designing protein sequences conditioned on a specific target fold. To model the complex sequence--structure relationship, Fold2Seq jointly learns a sequence embedding using a transformer and a fold embedding from the density of secondary structural elements in 3D voxels. On test sets with single, high-resolution and complete structure inputs for individual folds, our experiments demonstrate improved or comparable performance of Fold2Seq in terms of speed, coverage, and reliability for sequence design, when compared to existing state-of-the-art methods that include data-driven deep generative models and physics-based RosettaDesign. The unique advantages of fold-based Fold2Seq, in comparison to a structure-based deep model and RosettaDesign, become more evident on three additional real-world challenges originating from low-quality, incomplete, or ambiguous input structures. Source code and data are available at https://github.com/IBM/fold2seq.
Do Large Scale Molecular Language Representations Capture Important Structural Information?
Jun 17, 2021Jerret Ross, Brian Belgodere, Vijil Chenthamarakshan, Inkit Padhi, Youssef Mroueh, Payel Das
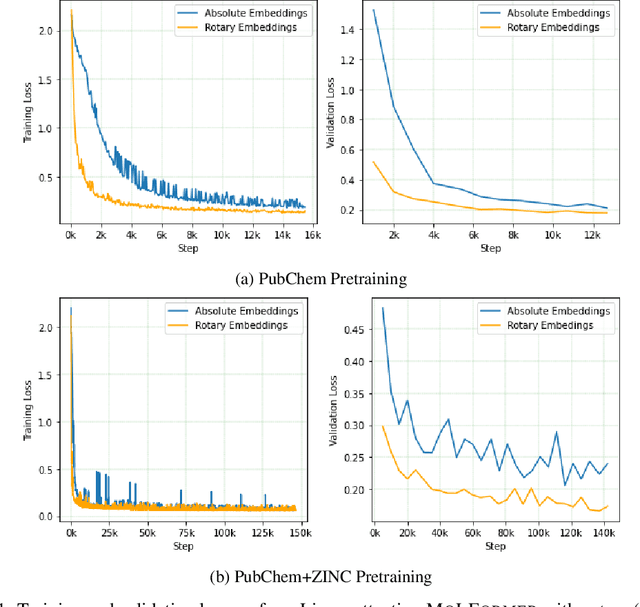
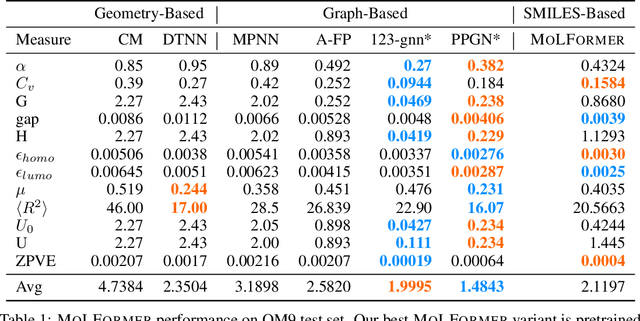
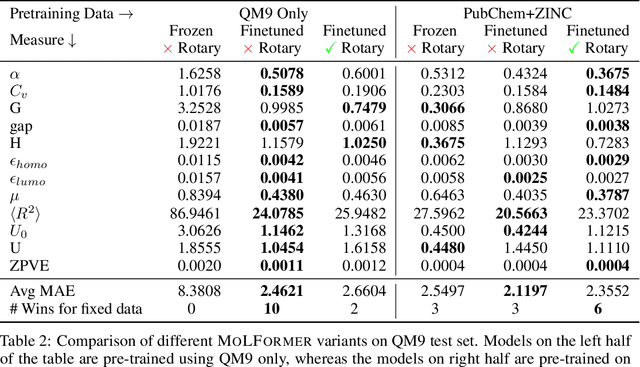
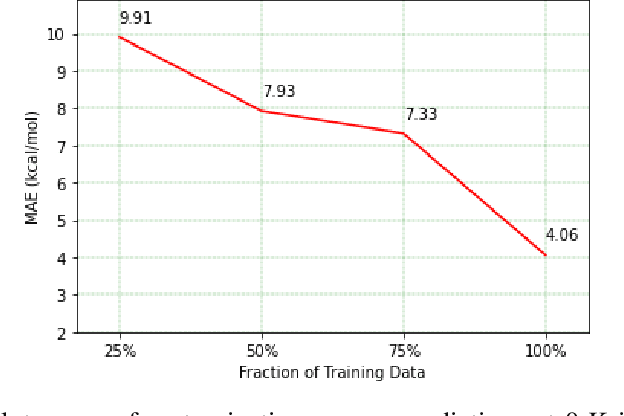
Predicting chemical properties from the structure of a molecule is of great importance in many applications including drug discovery and material design. Machine learning based molecular property prediction holds the promise of enabling accurate predictions at much less complexity, when compared to, for example Density Functional Theory (DFT) calculations. Features extracted from molecular graphs, using graph neural nets in a supervised manner, have emerged as strong baselines for such tasks. However, the vast chemical space together with the limited availability of labels makes supervised learning challenging, calling for learning a general-purpose molecular representation. Recently, pre-trained transformer-based language models (PTLMs) on large unlabeled corpus have produced state-of-the-art results in many downstream natural language processing tasks. Inspired by this development, here we present molecular embeddings obtained by training an efficient transformer encoder model, referred to as MoLFormer. This model was employed with a linear attention mechanism and highly paralleized training on 1D SMILES sequences of 1.1 billion unlabeled molecules from the PubChem and ZINC datasets. Experiments show that the learned molecular representation performs competitively, when compared to existing graph-based and fingerprint-based supervised learning baselines, on the challenging tasks of predicting properties of QM8 and QM9 molecules. Further task-specific fine-tuning of the MoLFormerr representation improves performance on several of those property prediction benchmarks. These results provide encouraging evidence that large-scale molecular language models can capture sufficient structural information to be able to accurately predict quantum chemical properties and beyond.
Augmenting Molecular Deep Generative Models with Topological Data Analysis Representations
Jun 08, 2021Yair Schiff, Vijil Chenthamarakshan, Samuel Hoffman, Karthikeyan Natesan Ramamurthy, Payel Das
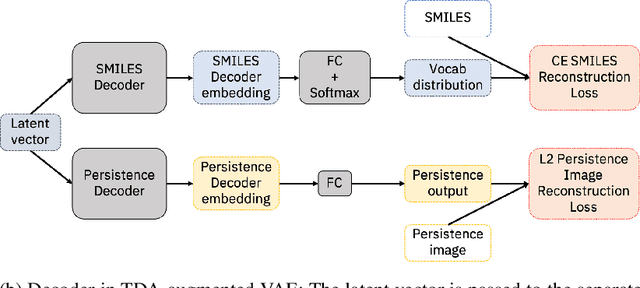

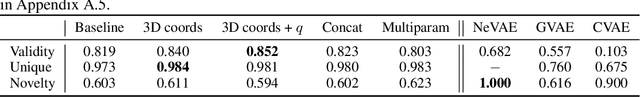
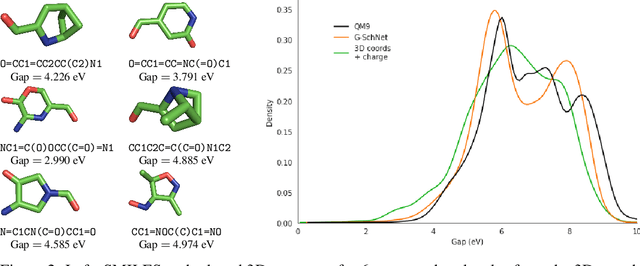
Deep generative models have emerged as a powerful tool for learning informative molecular representations and designing novel molecules with desired properties, with applications in drug discovery and material design. Deep generative auto-encoders defined over molecular SMILES strings have been a popular choice for that purpose. However, capturing salient molecular properties like quantum-chemical energies remains challenging and requires sophisticated neural net models of molecular graphs or geometry-based information. As a simpler and more efficient alternative, we present a SMILES Variational Auto-Encoder (VAE) augmented with topological data analysis (TDA) representations of molecules, known as persistence images. Our experiments show that this TDA augmentation enables a SMILES VAE to capture the complex relation between 3D geometry and electronic properties, and allows generation of novel, diverse, and valid molecules with geometric features consistent with the training data, which exhibit a varying range of global electronic structural properties, such as a small HOMO-LUMO gap - a critical property for designing organic solar cells. We demonstrate that our TDA augmentation yields better success in downstream tasks compared to models trained without these representations and can assist in targeted molecule discovery.
Optimizing Molecules using Efficient Queries from Property Evaluations
Nov 03, 2020Samuel Hoffman, Vijil Chenthamarakshan, Kahini Wadhawan, Pin-Yu Chen, Payel Das
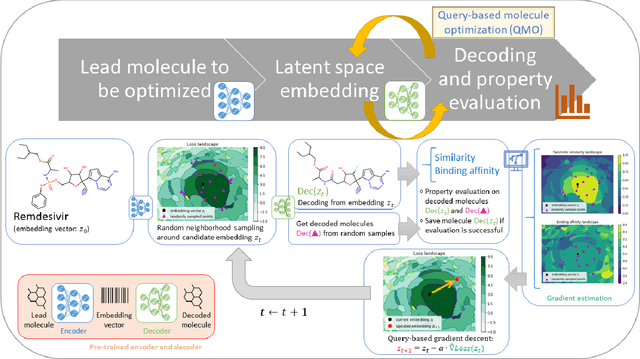
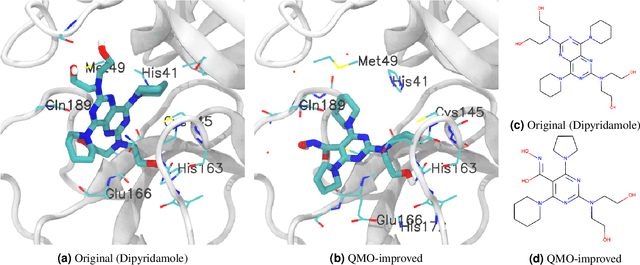
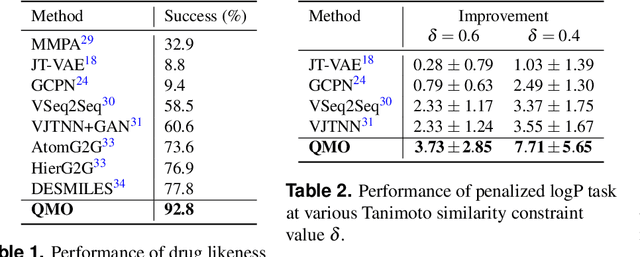
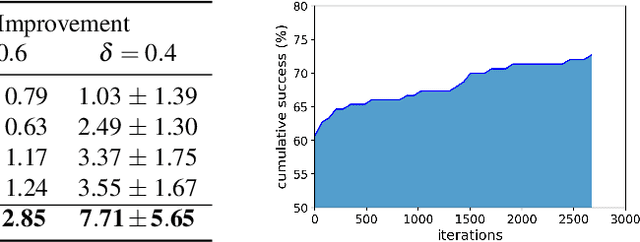
Machine learning has shown potential for optimizing existing molecules with more desirable properties, a critical step towards accelerating new chemical discovery. In this work, we propose QMO, a generic query-based molecule optimization framework that exploits latent embeddings from a molecule autoencoder. QMO improves the desired properties of an input molecule based on efficient queries, guided by a set of molecular property predictions and evaluation metrics. We show that QMO outperforms existing methods in the benchmark tasks of optimizing molecules for drug likeliness and solubility under similarity constraints. We also demonstrate significant property improvement using QMO on two new and challenging tasks that are also important in real-world discovery problems: (i) optimizing existing SARS-CoV-2 Main Protease inhibitors toward higher binding affinity; and (ii) improving known antimicrobial peptides towards lower toxicity. Results from QMO show high consistency with external validations, suggesting effective means of facilitating molecule optimization problems with design constraints.
Characterizing the Latent Space of Molecular Deep Generative Models with Persistent Homology Metrics
Oct 18, 2020Yair Schiff, Vijil Chenthamarakshan, Karthikeyan Natesan Ramamurthy, Payel Das
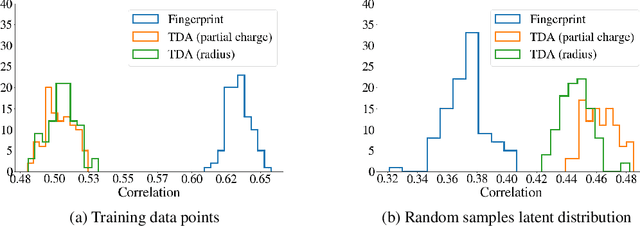
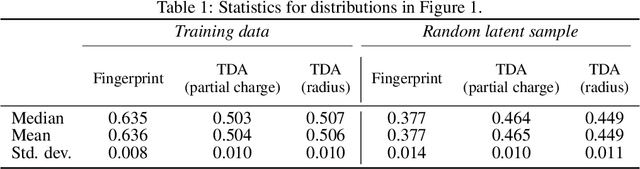
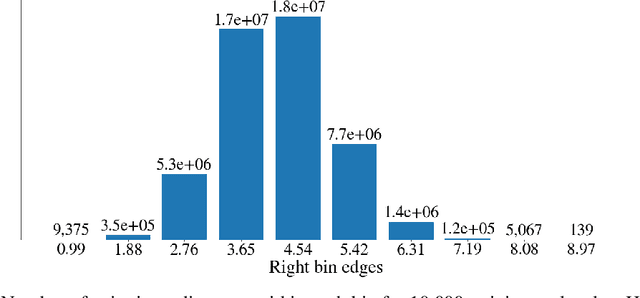
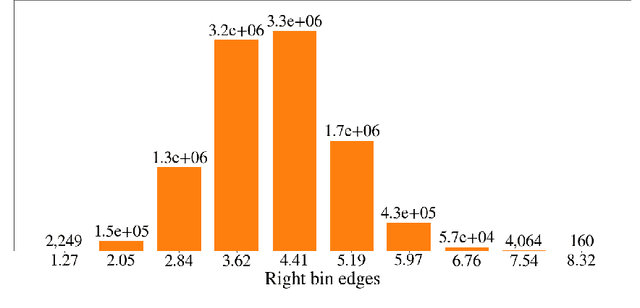
Deep generative models are increasingly becoming integral parts of the in silico molecule design pipeline and have dual goals of learning the chemical and structural features that render candidate molecules viable while also being flexible enough to generate novel designs. Specifically, Variational Auto Encoders (VAEs) are generative models in which encoder-decoder network pairs are trained to reconstruct training data distributions in such a way that the latent space of the encoder network is smooth. Therefore, novel candidates can be found by sampling from this latent space. However, the scope of architectures and hyperparameters is vast and choosing the best combination for in silico discovery has important implications for downstream success. Therefore, it is important to develop a principled methodology for distinguishing how well a given generative model is able to learn salient molecular features. In this work, we propose a method for measuring how well the latent space of deep generative models is able to encode structural and chemical features of molecular datasets by correlating latent space metrics with metrics from the field of topological data analysis (TDA). We apply our evaluation methodology to a VAE trained on SMILES strings and show that 3D topology information is consistently encoded throughout the latent space of the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge