Stefano Pedemonte
Whiterabbit AI, Inc., 3930 Freedom Circle Suite 101, Santa Clara, CA 95054
A deep learning algorithm for reducing false positives in screening mammography
Apr 13, 2022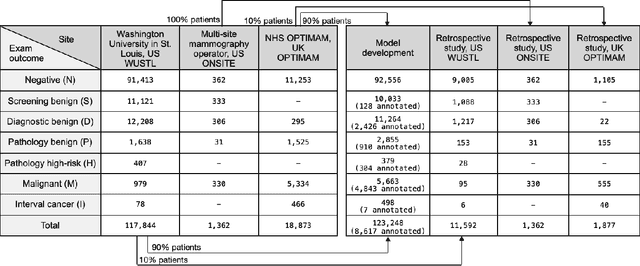
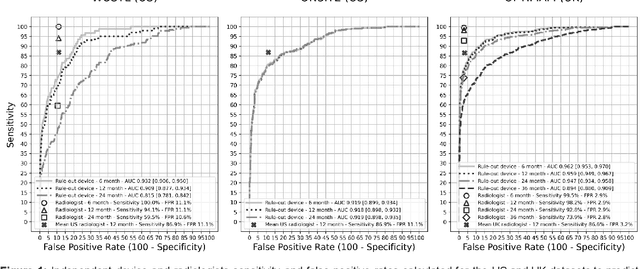
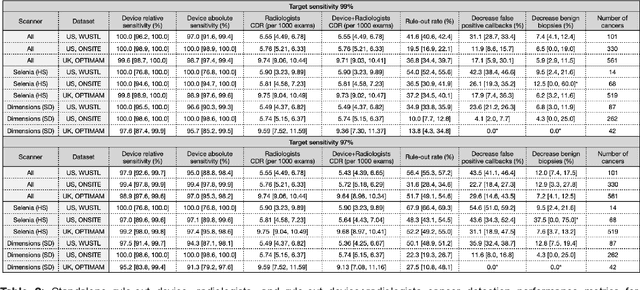
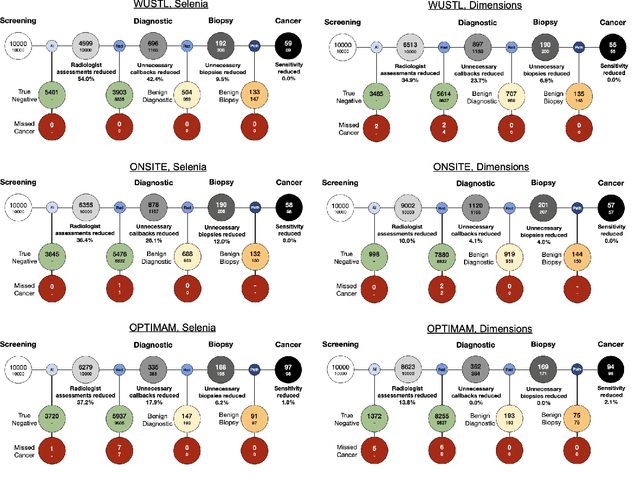
Abstract:Screening mammography improves breast cancer outcomes by enabling early detection and treatment. However, false positive callbacks for additional imaging from screening exams cause unnecessary procedures, patient anxiety, and financial burden. This work demonstrates an AI algorithm that reduces false positives by identifying mammograms not suspicious for breast cancer. We trained the algorithm to determine the absence of cancer using 123,248 2D digital mammograms (6,161 cancers) and performed a retrospective study on 14,831 screening exams (1,026 cancers) from 15 US and 3 UK sites. Retrospective evaluation of the algorithm on the largest of the US sites (11,592 mammograms, 101 cancers) a) left the cancer detection rate unaffected (p=0.02, non-inferiority margin 0.25 cancers per 1000 exams), b) reduced callbacks for diagnostic exams by 31.1% compared to standard clinical readings, c) reduced benign needle biopsies by 7.4%, and d) reduced screening exams requiring radiologist interpretation by 41.6% in the simulated clinical workflow. This work lays the foundation for semi-autonomous breast cancer screening systems that could benefit patients and healthcare systems by reducing false positives, unnecessary procedures, patient anxiety, and expenses.
A multi-site study of a breast density deep learning model for full-field digital mammography and digital breast tomosynthesis exams
Jan 23, 2020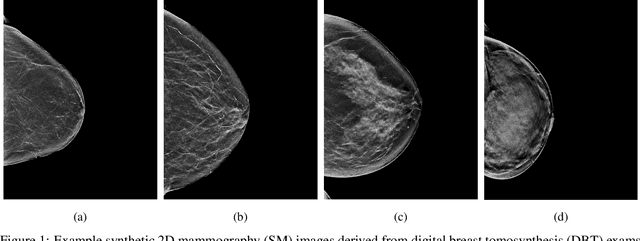
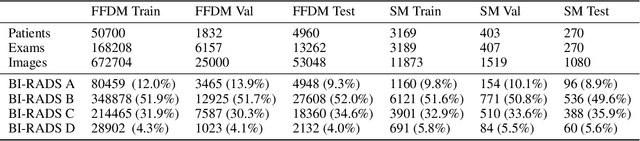
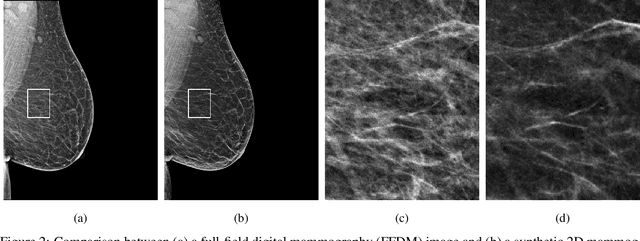
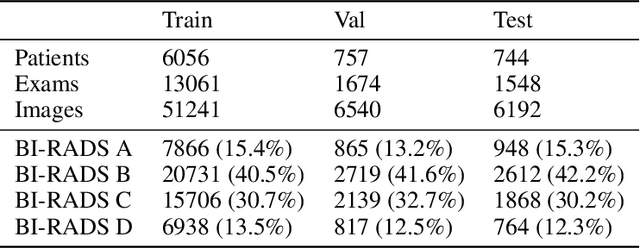
Abstract:$\textbf{Purpose:}$ To develop a Breast Imaging Reporting and Data System (BI-RADS) breast density DL model in a multi-site setting for synthetic 2D mammography (SM) images derived from 3D DBT exams using FFDM images and limited SM data. $\textbf{Materials and Methods:}$ A DL model was trained to predict BI-RADS breast density using FFDM images acquired from 2008 to 2017 (Site 1: 57492 patients, 187627 exams, 750752 images) for this retrospective study. The FFDM model was evaluated using SM datasets from two institutions (Site 1: 3842 patients, 3866 exams, 14472 images, acquired from 2016 to 2017; Site 2: 7557 patients, 16283 exams, 63973 images, 2015 to 2019). Adaptation methods were investigated to improve performance on the SM datasets and the effect of dataset size on each adaptation method is considered. Statistical significance was assessed using confidence intervals (CI), estimated by bootstrapping. $\textbf{Results:}$ Without adaptation, the model demonstrated close agreement with the original reporting radiologists for all three datasets (Site 1 FFDM: linearly-weighted $\kappa_w$ = 0.75, 95\% CI: [0.74, 0.76]; Site 1 SM: $\kappa_w$ = 0.71, CI: [0.64, 0.78]; Site 2 SM: $\kappa_w$ = 0.72, CI: [0.70, 0.75]). With adaptation, performance improved for Site 2 (Site 1: $\kappa_w$ = 0.72, CI: [0.66, 0.79], Site 2: $\kappa_w$ = 0.79, CI: [0.76, 0.81]) using only 500 SM images from each site. $\textbf{Conclusion:}$ A BI-RADS breast density DL model demonstrated strong performance on FFDM and SM images from two institutions without training on SM images and improved using few SM images.
A Hypersensitive Breast Cancer Detector
Jan 23, 2020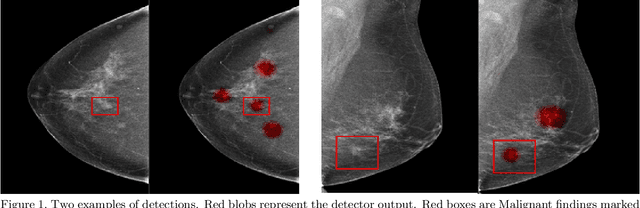
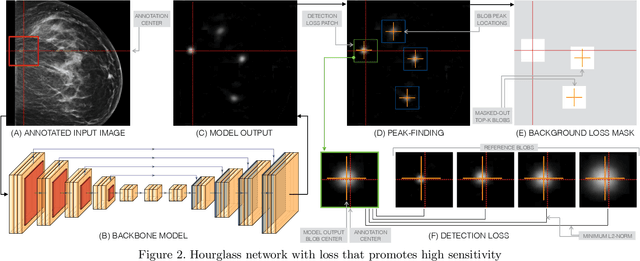
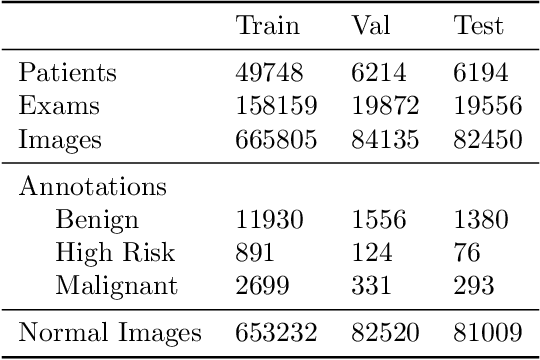
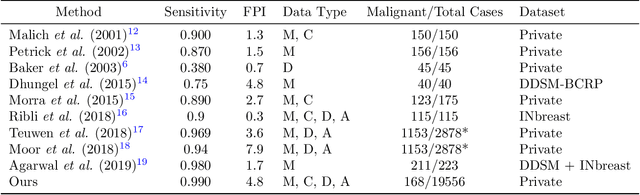
Abstract:Early detection of breast cancer through screening mammography yields a 20-35% increase in survival rate; however, there are not enough radiologists to serve the growing population of women seeking screening mammography. Although commercial computer aided detection (CADe) software has been available to radiologists for decades, it has failed to improve the interpretation of full-field digital mammography (FFDM) images due to its low sensitivity over the spectrum of findings. In this work, we leverage a large set of FFDM images with loose bounding boxes of mammographically significant findings to train a deep learning detector with extreme sensitivity. Building upon work from the Hourglass architecture, we train a model that produces segmentation-like images with high spatial resolution, with the aim of producing 2D Gaussian blobs centered on ground-truth boxes. We replace the pixel-wise $L_2$ norm with a weak-supervision loss designed to achieve high sensitivity, asymmetrically penalizing false positives and false negatives while softening the noise of the loose bounding boxes by permitting a tolerance in misaligned predictions. The resulting system achieves a sensitivity for malignant findings of 0.99 with only 4.8 false positive markers per image. When utilized in a CADe system, this model could enable a novel workflow where radiologists can focus their attention with trust on only the locations proposed by the model, expediting the interpretation process and bringing attention to potential findings that could otherwise have been missed. Due to its nearly perfect sensitivity, the proposed detector can also be used as a high-performance proposal generator in two-stage detection systems.
Adaptation of a deep learning malignancy model from full-field digital mammography to digital breast tomosynthesis
Jan 23, 2020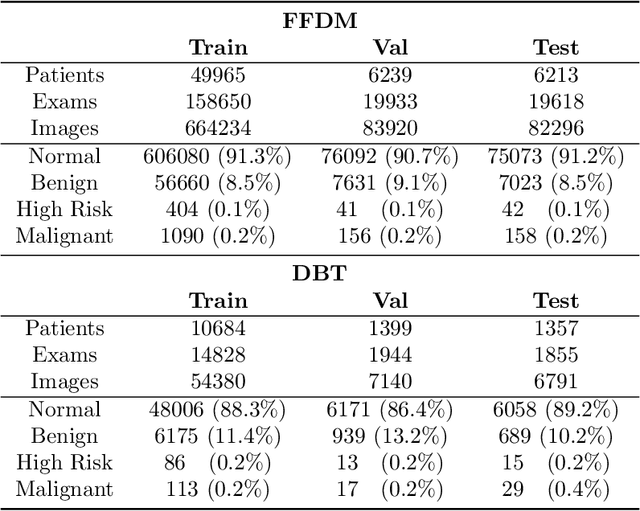
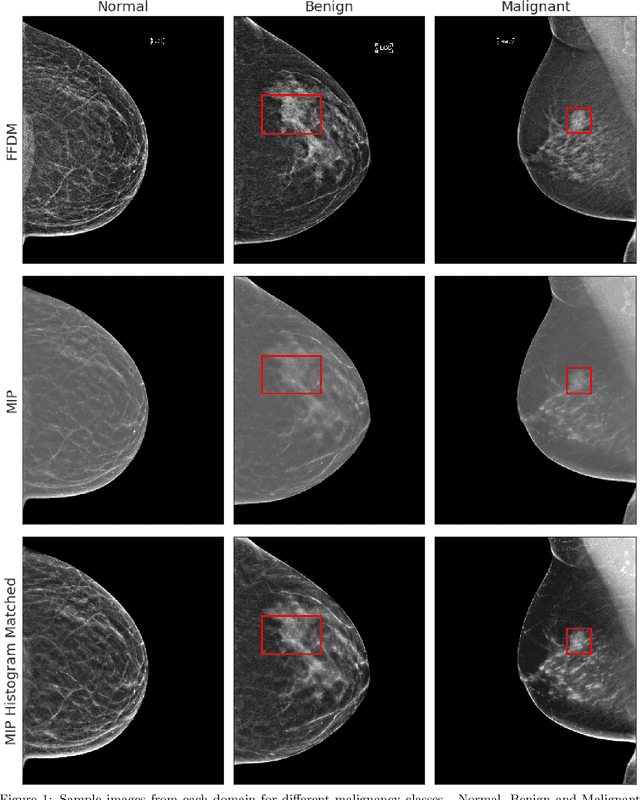
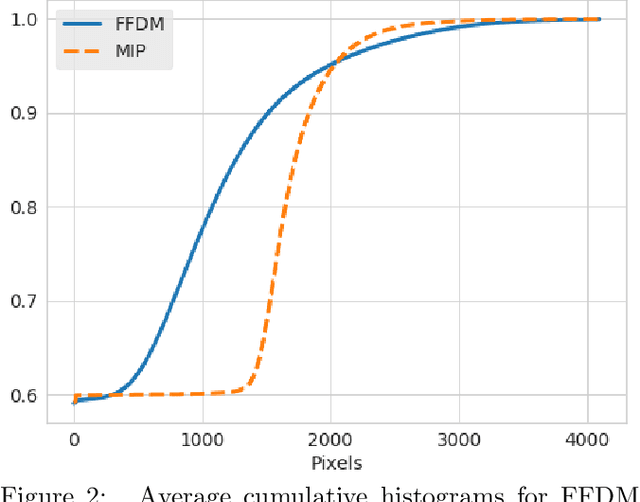
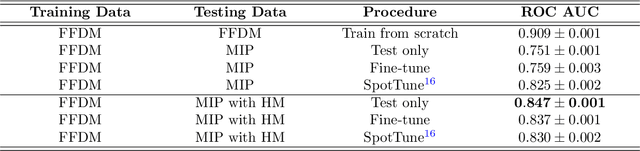
Abstract:Mammography-based screening has helped reduce the breast cancer mortality rate, but has also been associated with potential harms due to low specificity, leading to unnecessary exams or procedures, and low sensitivity. Digital breast tomosynthesis (DBT) improves on conventional mammography by increasing both sensitivity and specificity and is becoming common in clinical settings. However, deep learning (DL) models have been developed mainly on conventional 2D full-field digital mammography (FFDM) or scanned film images. Due to a lack of large annotated DBT datasets, it is difficult to train a model on DBT from scratch. In this work, we present methods to generalize a model trained on FFDM images to DBT images. In particular, we use average histogram matching (HM) and DL fine-tuning methods to generalize a FFDM model to the 2D maximum intensity projection (MIP) of DBT images. In the proposed approach, the differences between the FFDM and DBT domains are reduced via HM and then the base model, which was trained on abundant FFDM images, is fine-tuned. When evaluating on image patches extracted around identified findings, we are able to achieve similar areas under the receiver operating characteristic curve (ROC AUC) of $\sim 0.9$ for FFDM and $\sim 0.85$ for MIP images, as compared to a ROC AUC of $\sim 0.75$ when tested directly on MIP images.
Probabilistic Graphical Modeling approach to dynamic PET direct parametric map estimation and image reconstruction
Aug 24, 2018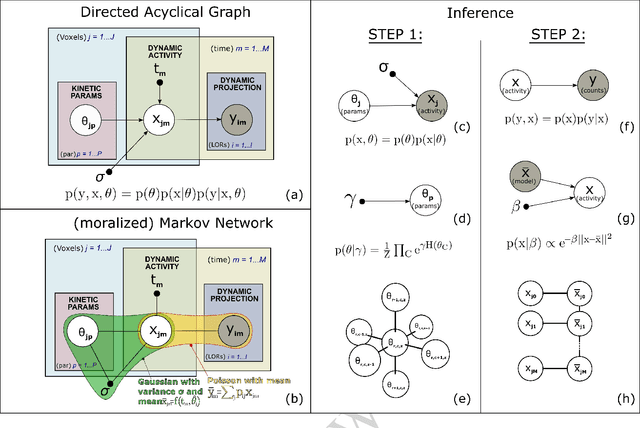
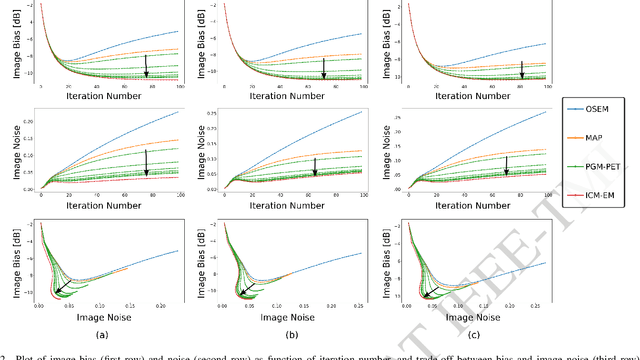
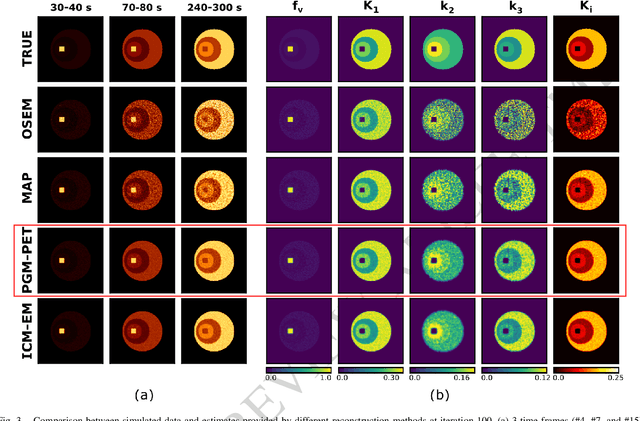
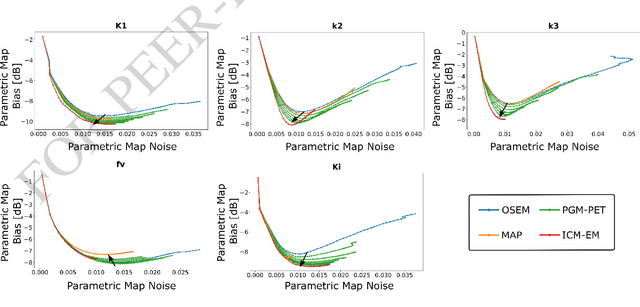
Abstract:In the context of dynamic emission tomography, the conventional processing pipeline consists of independent image reconstruction of single time frames, followed by the application of a suitable kinetic model to time activity curves (TACs) at the voxel or region-of-interest level. The relatively new field of 4D PET direct reconstruction, by contrast, seeks to move beyond this scheme and incorporate information from multiple time frames within the reconstruction task. Existing 4D direct models are based on a deterministic description of voxels' TACs, captured by the chosen kinetic model, considering the photon counting process the only source of uncertainty. In this work, we introduce a new probabilistic modeling strategy based on the key assumption that activity time course would be subject to uncertainty even if the parameters of the underlying dynamic process were known. This leads to a hierarchical Bayesian model, which we formulate using the formalism of Probabilistic Graphical Modeling (PGM). The inference of the joint probability density function arising from PGM is addressed using a new gradient-based iterative algorithm, which presents several advantages compared to existing direct methods: it is flexible to an arbitrary choice of linear and nonlinear kinetic model; it enables the inclusion of arbitrary (sub)differentiable priors for parametric maps; it is simpler to implement and suitable to integration in computing frameworks for machine learning. Computer simulations and an application to real patient scan showed how the proposed approach allows us to weight the importance of the kinetic model, providing a bridge between indirect and deterministic direct methods.
DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning
Jul 26, 2018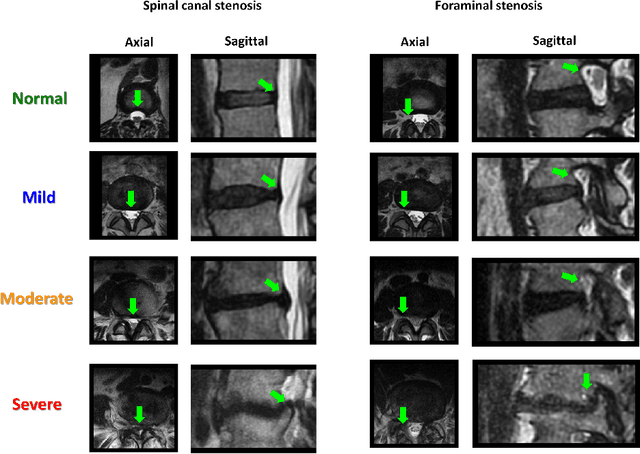
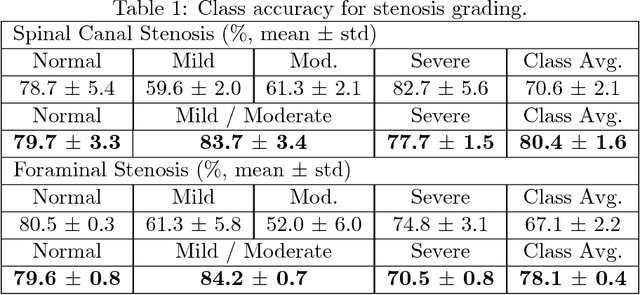
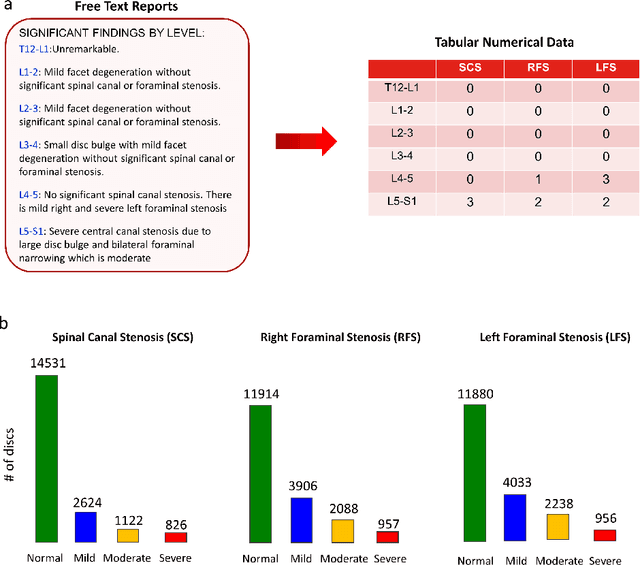
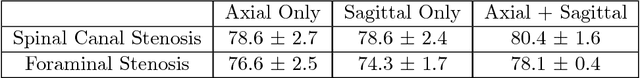
Abstract:The high prevalence of spinal stenosis results in a large volume of MRI imaging, yet interpretation can be time-consuming with high inter-reader variability even among the most specialized radiologists. In this paper, we develop an efficient methodology to leverage the subject-matter-expertise stored in large-scale archival reporting and image data for a deep-learning approach to fully-automated lumbar spinal stenosis grading. Specifically, we introduce three major contributions: (1) a natural-language-processing scheme to extract level-by-level ground-truth labels from free-text radiology reports for the various types and grades of spinal stenosis (2) accurate vertebral segmentation and disc-level localization using a U-Net architecture combined with a spine-curve fitting method, and (3) a multi-input, multi-task, and multi-class convolutional neural network to perform central canal and foraminal stenosis grading on both axial and sagittal imaging series inputs with the extracted report-derived labels applied to corresponding imaging level segments. This study uses a large dataset of 22796 disc-levels extracted from 4075 patients. We achieve state-of-the-art performance on lumbar spinal stenosis classification and expect the technique will increase both radiology workflow efficiency and the perceived value of radiology reports for referring clinicians and patients.
Kinetic Compressive Sensing
Mar 27, 2018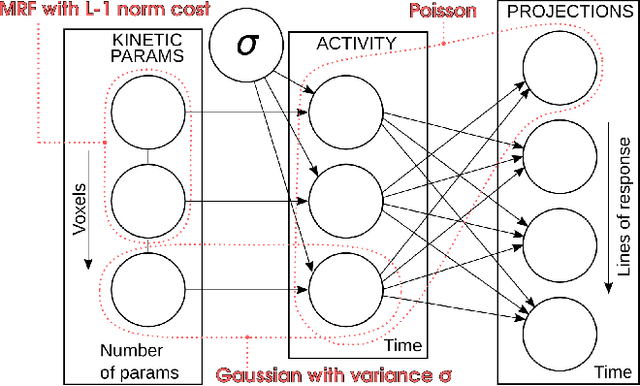
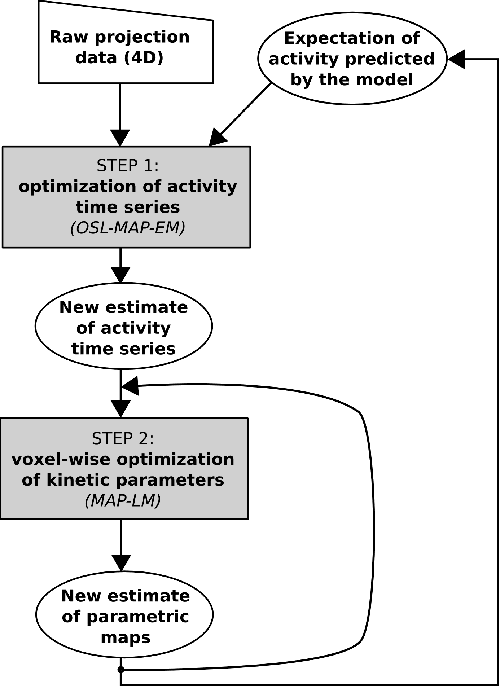
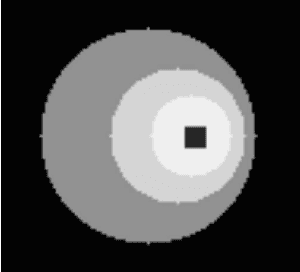

Abstract:Parametric images provide insight into the spatial distribution of physiological parameters, but they are often extremely noisy, due to low SNR of tomographic data. Direct estimation from projections allows accurate noise modeling, improving the results of post-reconstruction fitting. We propose a method, which we name kinetic compressive sensing (KCS), based on a hierarchical Bayesian model and on a novel reconstruction algorithm, that encodes sparsity of kinetic parameters. Parametric maps are reconstructed by maximizing the joint probability, with an Iterated Conditional Modes (ICM) approach, alternating the optimization of activity time series (OS-MAP-OSL), and kinetic parameters (MAP-LM). We evaluated the proposed algorithm on a simulated dynamic phantom: a bias/variance study confirmed how direct estimates can improve the quality of parametric maps over a post-reconstruction fitting, and showed how the novel sparsity prior can further reduce their variance, without affecting bias. Real FDG PET human brain data (Siemens mMR, 40min) images were also processed. Results enforced how the proposed KCS-regularized direct method can produce spatially coherent images and parametric maps, with lower spatial noise and better tissue contrast. A GPU-based open source implementation of the algorithm is provided.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge