Katherine P. Andriole
"Name that manufacturer". Relating image acquisition bias with task complexity when training deep learning models: experiments on head CT
Aug 19, 2020
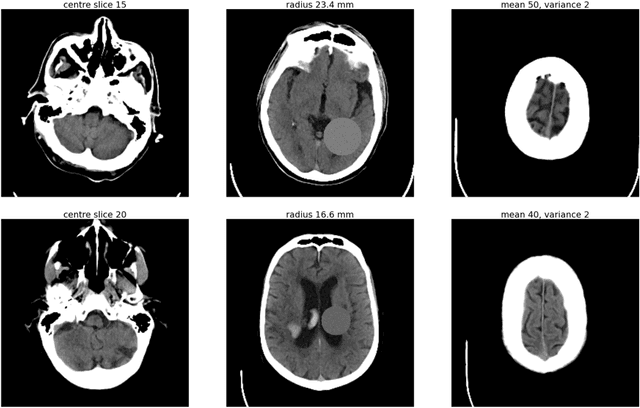

Abstract:As interest in applying machine learning techniques for medical images continues to grow at a rapid pace, models are starting to be developed and deployed for clinical applications. In the clinical AI model development lifecycle (described by Lu et al. [1]), a crucial phase for machine learning scientists and clinicians is the proper design and collection of the data cohort. The ability to recognize various forms of biases and distribution shifts in the dataset is critical at this step. While it remains difficult to account for all potential sources of bias, techniques can be developed to identify specific types of bias in order to mitigate their impact. In this work we analyze how the distribution of scanner manufacturers in a dataset can contribute to the overall bias of deep learning models. We evaluate convolutional neural networks (CNN) for both classification and segmentation tasks, specifically two state-of-the-art models: ResNet [2] for classification and U-Net [3] for segmentation. We demonstrate that CNNs can learn to distinguish the imaging scanner manufacturer and that this bias can substantially impact model performance for both classification and segmentation tasks. By creating an original synthesis dataset of brain data mimicking the presence of more or less subtle lesions we also show that this bias is related to the difficulty of the task. Recognition of such bias is critical to develop robust, generalizable models that will be crucial for clinical applications in real-world data distributions.
DeepAAA: clinically applicable and generalizable detection of abdominal aortic aneurysm using deep learning
Jul 04, 2019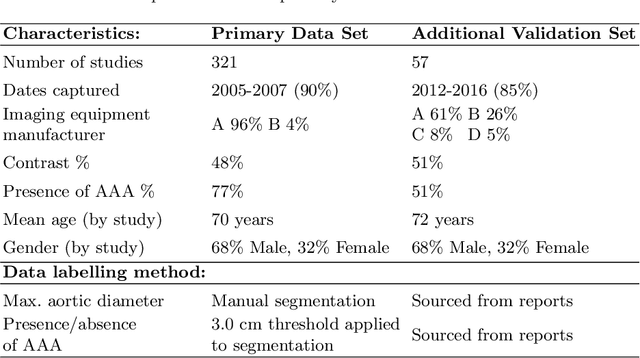
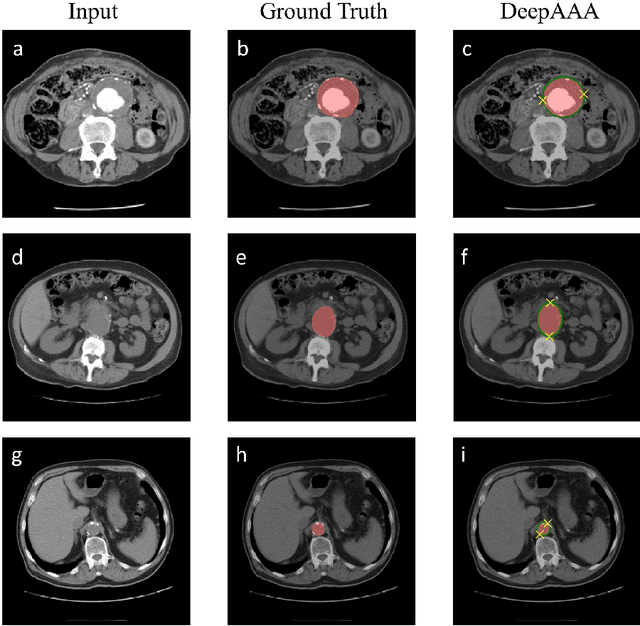
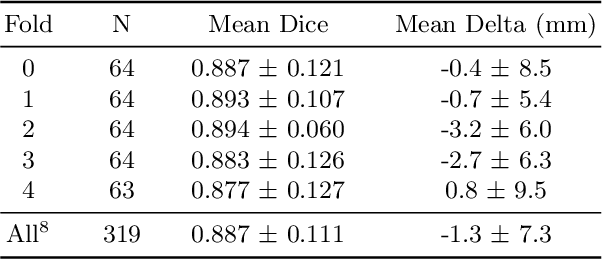
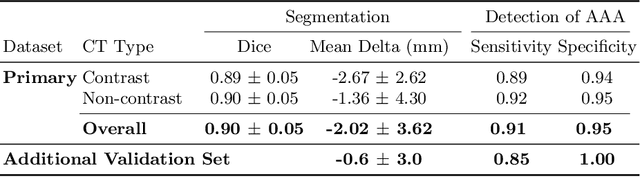
Abstract:We propose a deep learning-based technique for detection and quantification of abdominal aortic aneurysms (AAAs). The condition, which leads to more than 10,000 deaths per year in the United States, is asymptomatic, often detected incidentally, and often missed by radiologists. Our model architecture is a modified 3D U-Net combined with ellipse fitting that performs aorta segmentation and AAA detection. The study uses 321 abdominal-pelvic CT examinations performed by Massachusetts General Hospital Department of Radiology for training and validation. The model is then further tested for generalizability on a separate set of 57 examinations with differing patient demographics and acquisition characteristics than the original dataset. DeepAAA achieves high performance on both sets of data (sensitivity/specificity 0.91/0.95 and 0.85 / 1.0 respectively), on contrast and non-contrast CT scans and works with image volumes with varying numbers of images. We find that DeepAAA exceeds literature-reported performance of radiologists on incidental AAA detection. It is expected that the model can serve as an effective background detector in routine CT examinations to prevent incidental AAAs from being missed.
DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning
Jul 26, 2018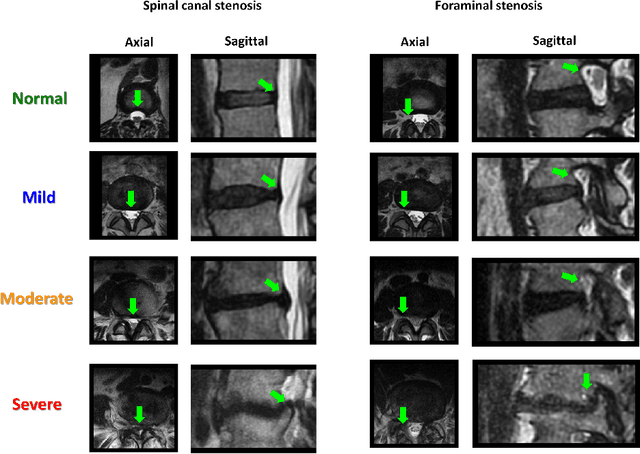
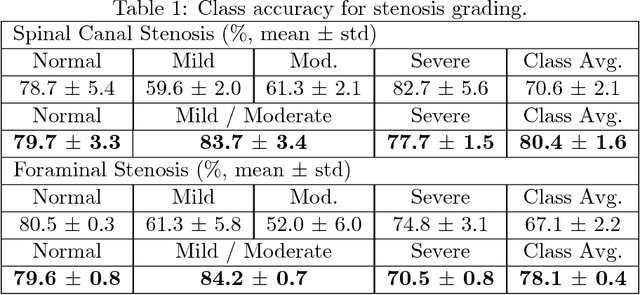
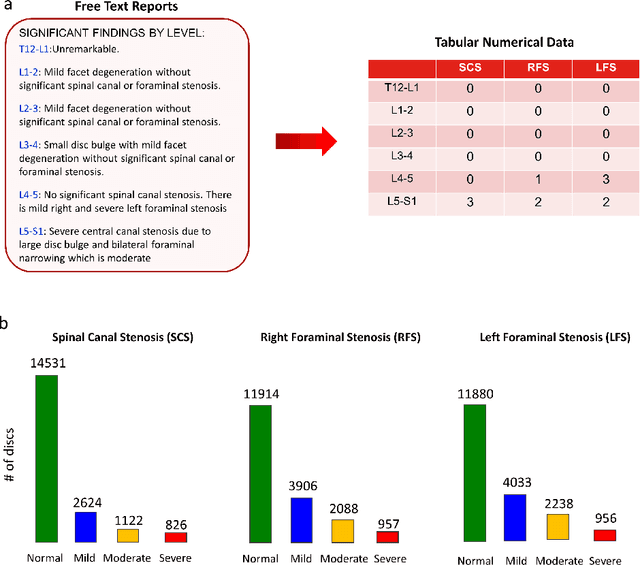
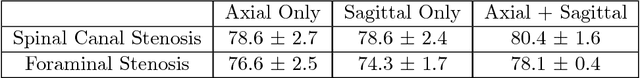
Abstract:The high prevalence of spinal stenosis results in a large volume of MRI imaging, yet interpretation can be time-consuming with high inter-reader variability even among the most specialized radiologists. In this paper, we develop an efficient methodology to leverage the subject-matter-expertise stored in large-scale archival reporting and image data for a deep-learning approach to fully-automated lumbar spinal stenosis grading. Specifically, we introduce three major contributions: (1) a natural-language-processing scheme to extract level-by-level ground-truth labels from free-text radiology reports for the various types and grades of spinal stenosis (2) accurate vertebral segmentation and disc-level localization using a U-Net architecture combined with a spine-curve fitting method, and (3) a multi-input, multi-task, and multi-class convolutional neural network to perform central canal and foraminal stenosis grading on both axial and sagittal imaging series inputs with the extracted report-derived labels applied to corresponding imaging level segments. This study uses a large dataset of 22796 disc-levels extracted from 4075 patients. We achieve state-of-the-art performance on lumbar spinal stenosis classification and expect the technique will increase both radiology workflow efficiency and the perceived value of radiology reports for referring clinicians and patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge