Pedro Costa
ExplainFix: Explainable Spatially Fixed Deep Networks
Mar 18, 2023Abstract:Is there an initialization for deep networks that requires no learning? ExplainFix adopts two design principles: the "fixed filters" principle that all spatial filter weights of convolutional neural networks can be fixed at initialization and never learned, and the "nimbleness" principle that only few network parameters suffice. We contribute (a) visual model-based explanations, (b) speed and accuracy gains, and (c) novel tools for deep convolutional neural networks. ExplainFix gives key insights that spatially fixed networks should have a steered initialization, that spatial convolution layers tend to prioritize low frequencies, and that most network parameters are not necessary in spatially fixed models. ExplainFix models have up to 100x fewer spatial filter kernels than fully learned models and matching or improved accuracy. Our extensive empirical analysis confirms that ExplainFix guarantees nimbler models (train up to 17\% faster with channel pruning), matching or improved predictive performance (spanning 13 distinct baseline models, four architectures and two medical image datasets), improved robustness to larger learning rate, and robustness to varying model size. We are first to demonstrate that all spatial filters in state-of-the-art convolutional deep networks can be fixed at initialization, not learned.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Colon Nuclei Instance Segmentation using a Probabilistic Two-Stage Detector
Mar 01, 2022
Abstract:Cancer is one of the leading causes of death in the developed world. Cancer diagnosis is performed through the microscopic analysis of a sample of suspicious tissue. This process is time consuming and error prone, but Deep Learning models could be helpful for pathologists during cancer diagnosis. We propose to change the CenterNet2 object detection model to also perform instance segmentation, which we call SegCenterNet2. We train SegCenterNet2 in the CoNIC challenge dataset and show that it performs better than Mask R-CNN in the competition metrics.
AutoFITS: Automatic Feature Engineering for Irregular Time Series
Dec 29, 2021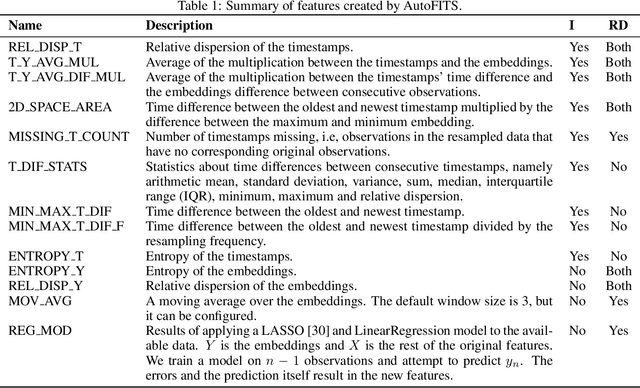
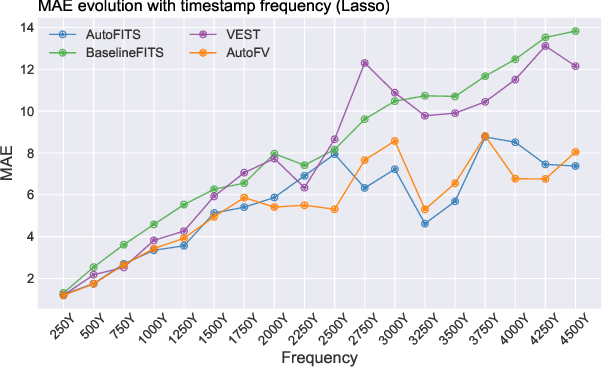
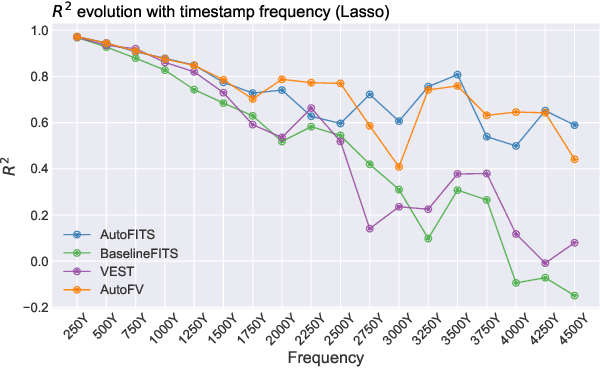
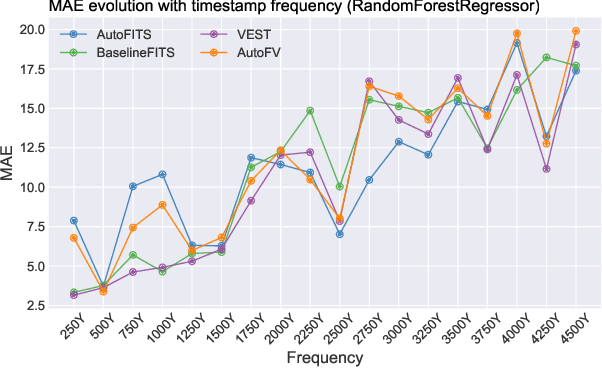
Abstract:A time series represents a set of observations collected over time. Typically, these observations are captured with a uniform sampling frequency (e.g. daily). When data points are observed in uneven time intervals the time series is referred to as irregular or intermittent. In such scenarios, the most common solution is to reconstruct the time series to make it regular, thus removing its intermittency. We hypothesise that, in irregular time series, the time at which each observation is collected may be helpful to summarise the dynamics of the data and improve forecasting performance. We study this idea by developing a novel automatic feature engineering framework, which focuses on extracting information from this point of view, i.e., when each instance is collected. We study how valuable this information is by integrating it in a time series forecasting workflow and investigate how it compares to or complements state-of-the-art methods for regular time series forecasting. In the end, we contribute by providing a novel framework that tackles feature engineering for time series from an angle previously vastly ignored. We show that our approach has the potential to further extract more information about time series that significantly improves forecasting performance.
Improving Lesion Segmentation for Diabetic Retinopathy using Adversarial Learning
Jul 27, 2020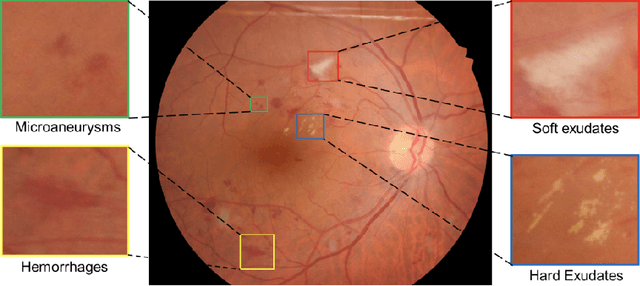
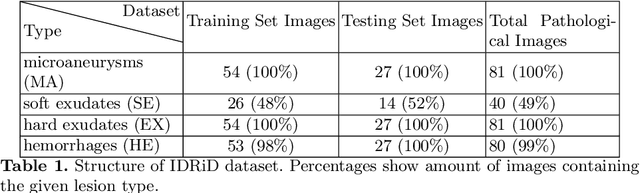
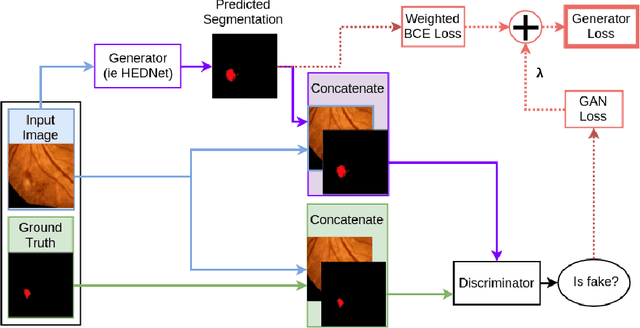
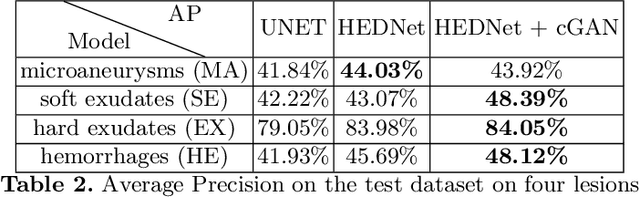
Abstract:Diabetic Retinopathy (DR) is a leading cause of blindness in working age adults. DR lesions can be challenging to identify in fundus images, and automatic DR detection systems can offer strong clinical value. Of the publicly available labeled datasets for DR, the Indian Diabetic Retinopathy Image Dataset (IDRiD) presents retinal fundus images with pixel-level annotations of four distinct lesions: microaneurysms, hemorrhages, soft exudates and hard exudates. We utilize the HEDNet edge detector to solve a semantic segmentation task on this dataset, and then propose an end-to-end system for pixel-level segmentation of DR lesions by incorporating HEDNet into a Conditional Generative Adversarial Network (cGAN). We design a loss function that adds adversarial loss to segmentation loss. Our experiments show that the addition of the adversarial loss improves the lesion segmentation performance over the baseline.
Learned Pre-Processing for Automatic Diabetic Retinopathy Detection on Eye Fundus Images
Jul 27, 2020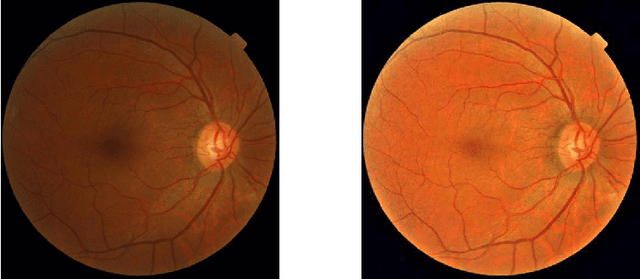
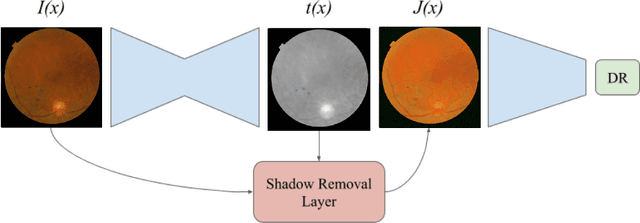
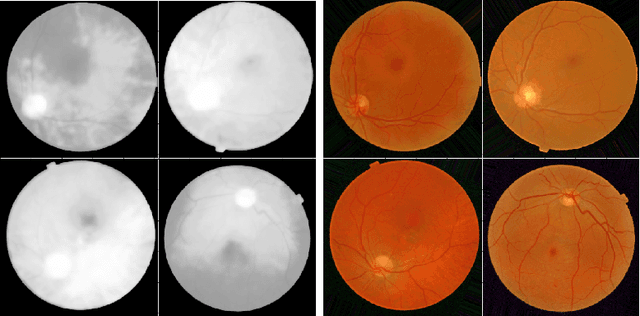
Abstract:Diabetic Retinopathy is the leading cause of blindness in the working-age population of the world. The main aim of this paper is to improve the accuracy of Diabetic Retinopathy detection by implementing a shadow removal and color correction step as a preprocessing stage from eye fundus images. For this, we rely on recent findings indicating that application of image dehazing on the inverted intensity domain amounts to illumination compensation. Inspired by this work, we propose a Shadow Removal Layer that allows us to learn the pre-processing function for a particular task. We show that learning the pre-processing function improves the performance of the network on the Diabetic Retinopathy detection task.
O-MedAL: Online Active Deep Learning for Medical Image Analysis
Aug 28, 2019
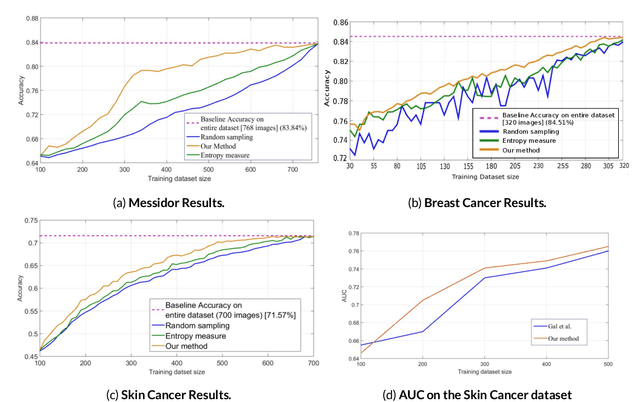
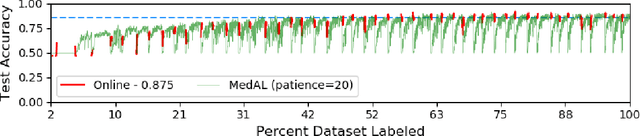
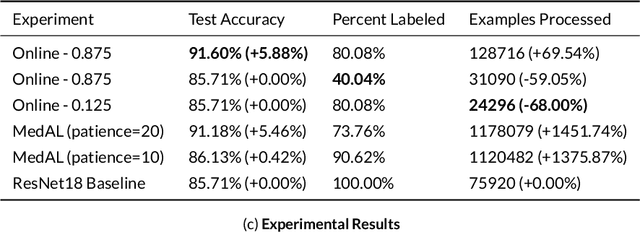
Abstract:Active Learning methods create an optimized and labeled training set from unlabeled data. We introduce a novel Online Active Deep Learning method for Medical Image Analysis. We extend our MedAL active learning framework to present new results in this paper. Experiments on three medical image datasets show that our novel online active learning model requires significantly less labelings, is more accurate, and is more robust to class imbalances than existing methods. Our method is also more accurate and computationally efficient than the baseline model. Compared to random sampling and uncertainty sampling, the method uses 275 and 200 (out of 768) fewer labeled examples, respectively. For Diabetic Retinopathy detection, our method attains a 5.88% accuracy improvement over the baseline model when 80% of the dataset is labeled, and the model reaches baseline accuracy when only 40% is labeled.
UOLO - automatic object detection and segmentation in biomedical images
Oct 09, 2018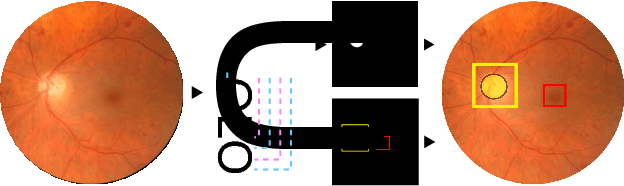
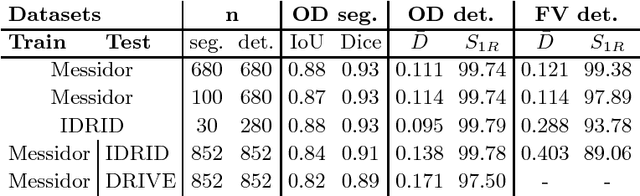
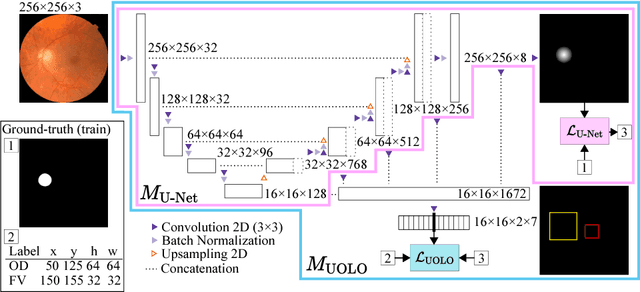
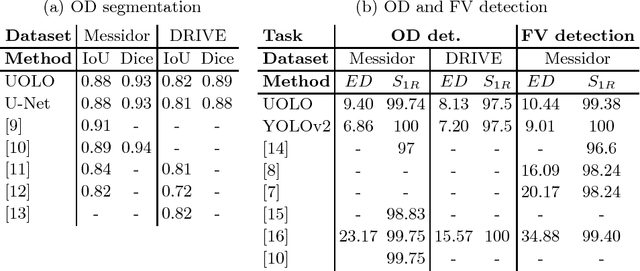
Abstract:We propose UOLO, a novel framework for the simultaneous detection and segmentation of structures of interest in medical images. UOLO consists of an object segmentation module which intermediate abstract representations are processed and used as input for object detection. The resulting system is optimized simultaneously for detecting a class of objects and segmenting an optionally different class of structures. UOLO is trained on a set of bounding boxes enclosing the objects to detect, as well as pixel-wise segmentation information, when available. A new loss function is devised, taking into account whether a reference segmentation is accessible for each training image, in order to suitably backpropagate the error. We validate UOLO on the task of simultaneous optic disc (OD) detection, fovea detection, and OD segmentation from retinal images, achieving state-of-the-art performance on public datasets.
* Publised on DLMIA 2018. Licensed under the Creative Commons CC-BY-NC-ND 4.0 license: http://creativecommons.org/licenses/by-nc-nd/4.0/
MedAL: Deep Active Learning Sampling Method for Medical Image Analysis
Sep 28, 2018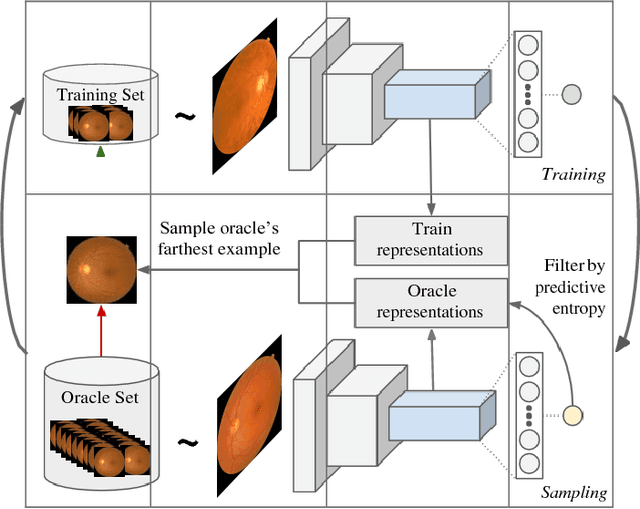
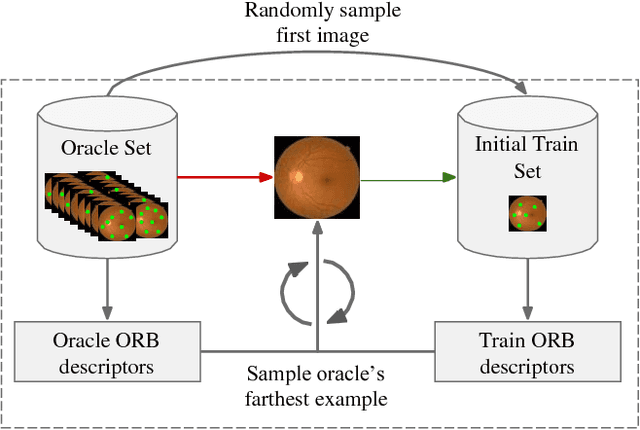
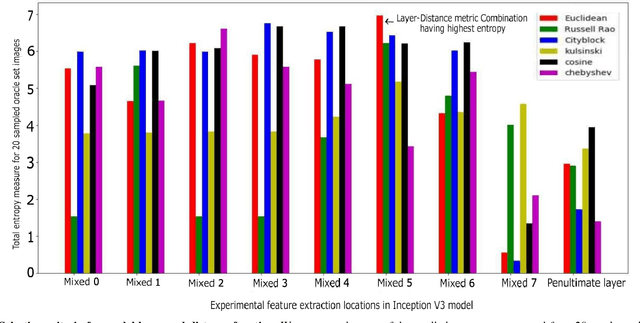
Abstract:Deep learning models have been successfully used in medical image analysis problems but they require a large amount of labeled images to obtain good performance.Deep learning models have been successfully used in medical image analysis problems but they require a large amount of labeled images to obtain good performance. However, such large labeled datasets are costly to acquire. Active learning techniques can be used to minimize the number of required training labels while maximizing the model's performance.In this work, we propose a novel sampling method that queries the unlabeled examples that maximize the average distance to all training set examples in a learned feature space. We then extend our sampling method to define a better initial training set, without the need for a trained model, by using ORB feature descriptors. We validate MedAL on 3 medical image datasets and show that our method is robust to different dataset properties. MedAL is also efficient, achieving 80% accuracy on the task of Diabetic Retinopathy detection using only 425 labeled images, corresponding to a 32% reduction in the number of required labeled examples compared to the standard uncertainty sampling technique, and a 40% reduction compared to random sampling.
Data-Driven Color Augmentation Techniques for Deep Skin Image Analysis
Mar 10, 2017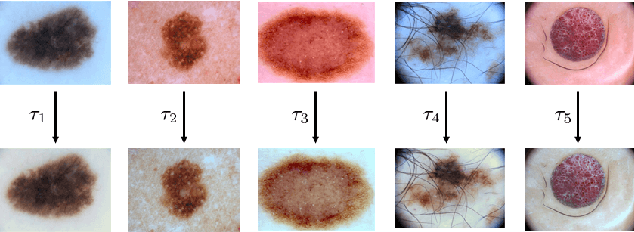
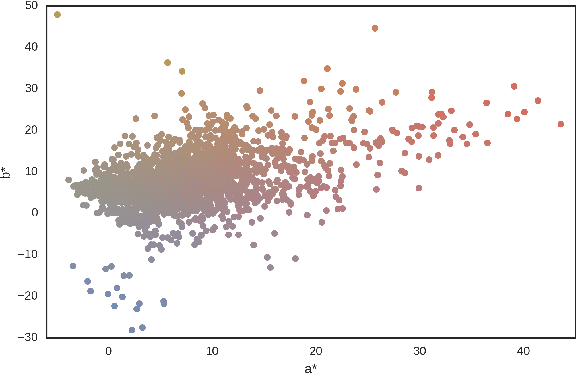
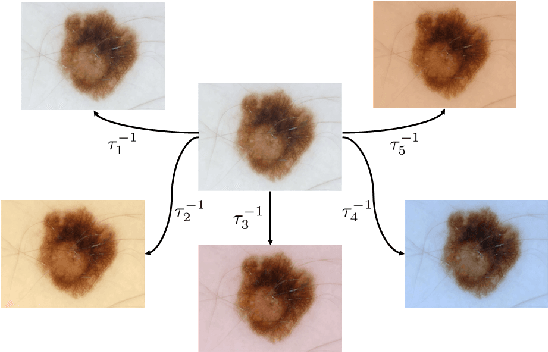
Abstract:Dermoscopic skin images are often obtained with different imaging devices, under varying acquisition conditions. In this work, instead of attempting to perform intensity and color normalization, we propose to leverage computational color constancy techniques to build an artificial data augmentation technique suitable for this kind of images. Specifically, we apply the \emph{shades of gray} color constancy technique to color-normalize the entire training set of images, while retaining the estimated illuminants. We then draw one sample from the distribution of training set illuminants and apply it on the normalized image. We employ this technique for training two deep convolutional neural networks for the tasks of skin lesion segmentation and skin lesion classification, in the context of the ISIC 2017 challenge and without using any external dermatologic image set. Our results on the validation set are promising, and will be supplemented with extended results on the hidden test set when available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge