Aurélio Campilho
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
HeartSpot: Privatized and Explainable Data Compression for Cardiomegaly Detection
Oct 05, 2022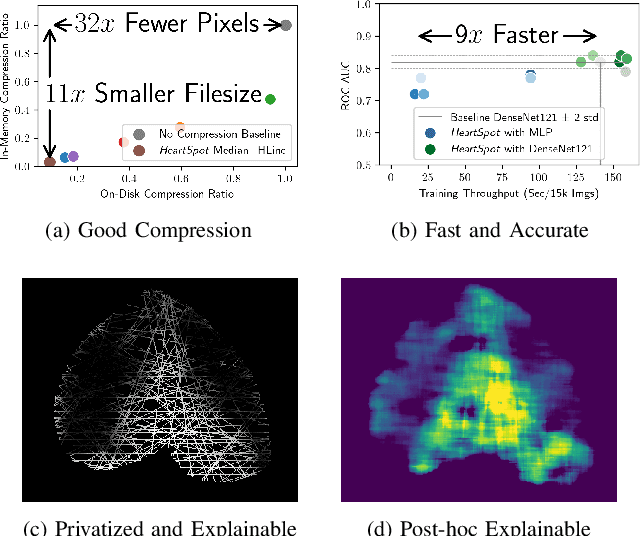
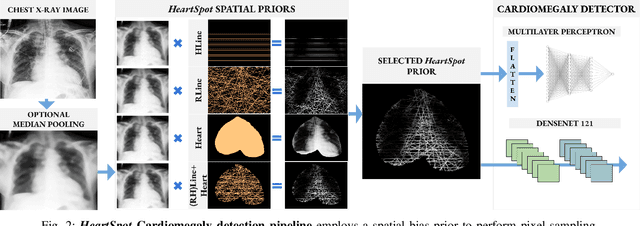
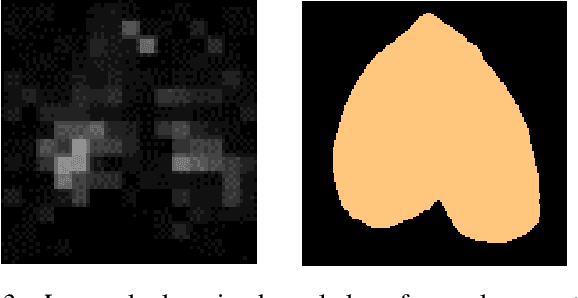
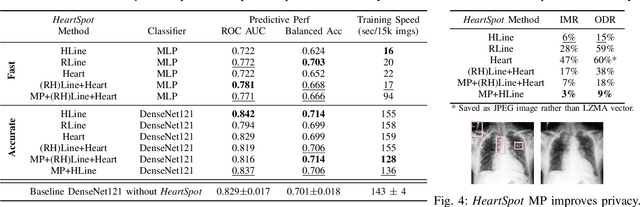
Abstract:Advances in data-driven deep learning for chest X-ray image analysis underscore the need for explainability, privacy, large datasets and significant computational resources. We frame privacy and explainability as a lossy single-image compression problem to reduce both computational and data requirements without training. For Cardiomegaly detection in chest X-ray images, we propose HeartSpot and four spatial bias priors. HeartSpot priors define how to sample pixels based on domain knowledge from medical literature and from machines. HeartSpot privatizes chest X-ray images by discarding up to 97% of pixels, such as those that reveal the shape of the thoracic cage, bones, small lesions and other sensitive features. HeartSpot priors are ante-hoc explainable and give a human-interpretable image of the preserved spatial features that clearly outlines the heart. HeartSpot offers strong compression, with up to 32x fewer pixels and 11x smaller filesize. Cardiomegaly detectors using HeartSpot are up to 9x faster to train or at least as accurate (up to +.01 AUC ROC) when compared to a baseline DenseNet121. HeartSpot is post-hoc explainable by re-using existing attribution methods without requiring access to the original non-privatized image. In summary, HeartSpot improves speed and accuracy, reduces image size, improves privacy and ensures explainability. Source code: https://www.github.com/adgaudio/HeartSpot
Colon Nuclei Instance Segmentation using a Probabilistic Two-Stage Detector
Mar 01, 2022
Abstract:Cancer is one of the leading causes of death in the developed world. Cancer diagnosis is performed through the microscopic analysis of a sample of suspicious tissue. This process is time consuming and error prone, but Deep Learning models could be helpful for pathologists during cancer diagnosis. We propose to change the CenterNet2 object detection model to also perform instance segmentation, which we call SegCenterNet2. We train SegCenterNet2 in the CoNIC challenge dataset and show that it performs better than Mask R-CNN in the competition metrics.
Enhancement of Retinal Fundus Images via Pixel Color Amplification
Jul 28, 2020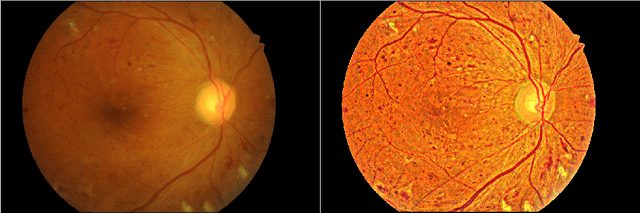
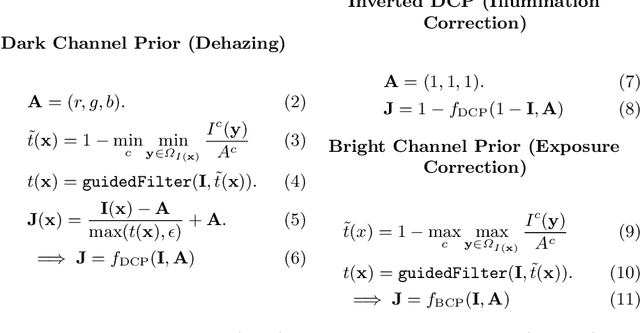
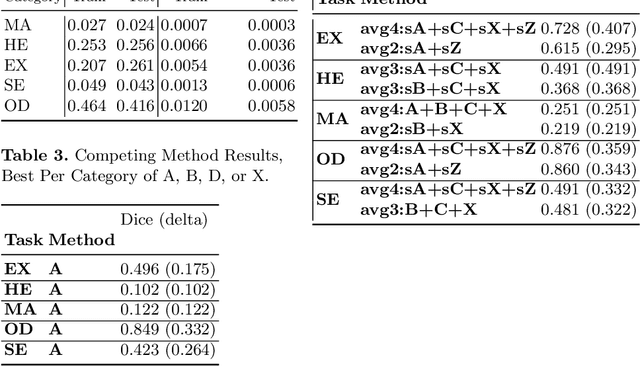
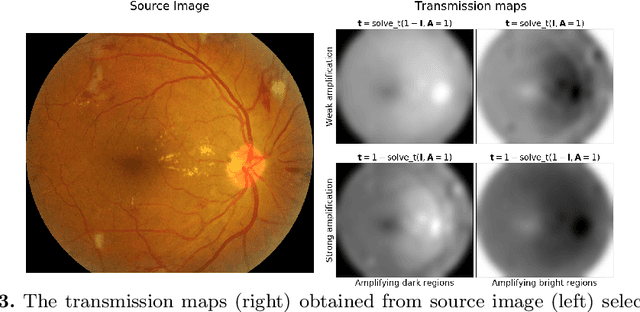
Abstract:We propose a pixel color amplification theory and family of enhancement methods to facilitate segmentation tasks on retinal images. Our novel re-interpretation of the image distortion model underlying dehazing theory shows how three existing priors commonly used by the dehazing community and a novel fourth prior are related. We utilize the theory to develop a family of enhancement methods for retinal images, including novel methods for whole image brightening and darkening. We show a novel derivation of the Unsharp Masking algorithm. We evaluate the enhancement methods as a pre-processing step to a challenging multi-task segmentation problem and show large increases in performance on all tasks, with Dice score increases over a no-enhancement baseline by as much as 0.491. We provide evidence that our enhancement preprocessing is useful for unbalanced and difficult data. We show that the enhancements can perform class balancing by composing them together.
Learned Pre-Processing for Automatic Diabetic Retinopathy Detection on Eye Fundus Images
Jul 27, 2020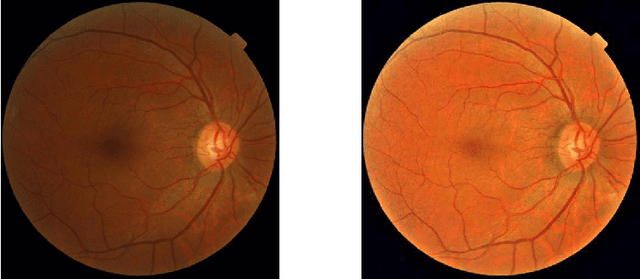
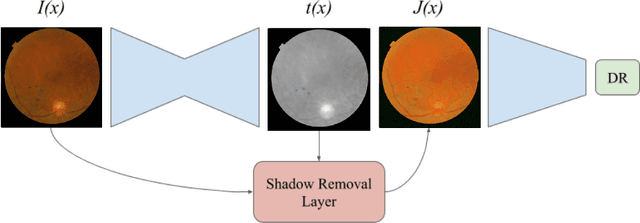
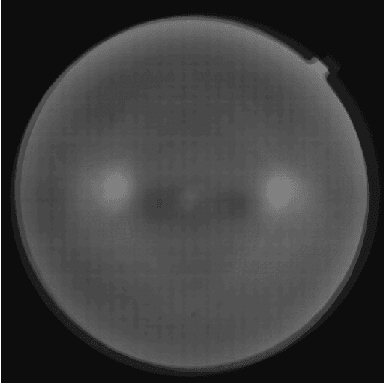
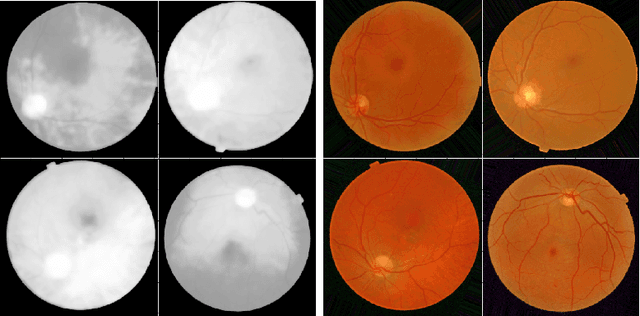
Abstract:Diabetic Retinopathy is the leading cause of blindness in the working-age population of the world. The main aim of this paper is to improve the accuracy of Diabetic Retinopathy detection by implementing a shadow removal and color correction step as a preprocessing stage from eye fundus images. For this, we rely on recent findings indicating that application of image dehazing on the inverted intensity domain amounts to illumination compensation. Inspired by this work, we propose a Shadow Removal Layer that allows us to learn the pre-processing function for a particular task. We show that learning the pre-processing function improves the performance of the network on the Diabetic Retinopathy detection task.
LNDb: A Lung Nodule Database on Computed Tomography
Dec 19, 2019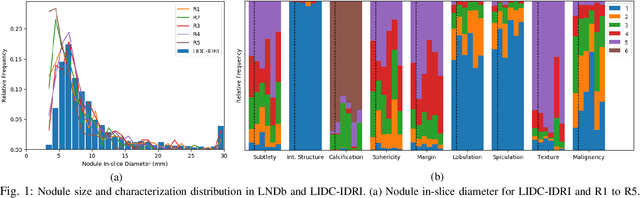
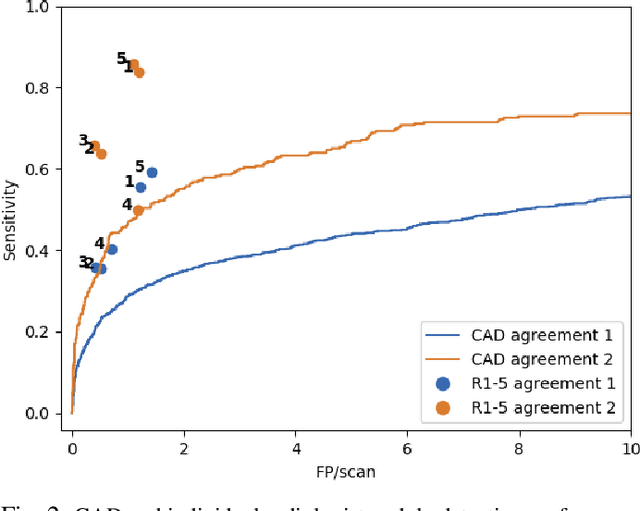
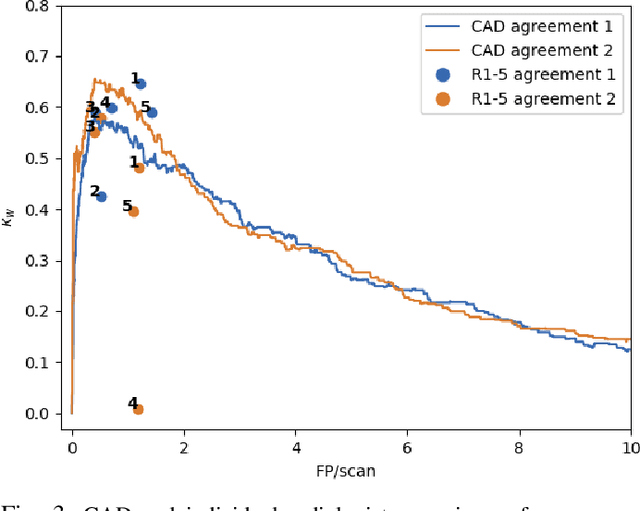
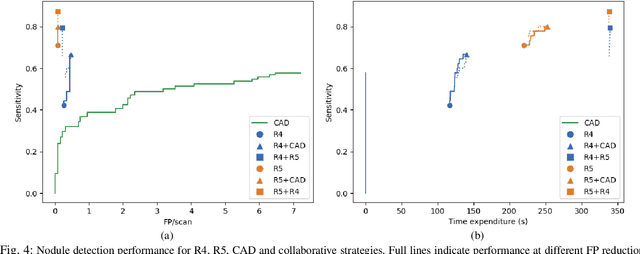
Abstract:Lung cancer is the deadliest type of cancer worldwide and late detection is the major factor for the low survival rate of patients. Low dose computed tomography has been suggested as a potential screening tool but manual screening is costly, time-consuming and prone to variability. This has fueled the development of automatic methods for the detection, segmentation and characterisation of pulmonary nodules but its application to clinical routine is challenging. In this study, a new database for the development and testing of pulmonary nodule computer-aided strategies is presented which intends to complement current databases by giving additional focus to radiologist variability and local clinical reality. State-of-the-art nodule detection, segmentation and characterization methods are tested and compared to manual annotations as well as collaborative strategies combining multiple radiologists and radiologists and computer-aided systems. It is shown that state-of-the-art methodologies can determine a patient's follow-up recommendation as accurately as a radiologist, though the nodule detection method used shows decreased performance in this database.
DR$\vert$GRADUATE: uncertainty-aware deep learning-based diabetic retinopathy grading in eye fundus images
Oct 25, 2019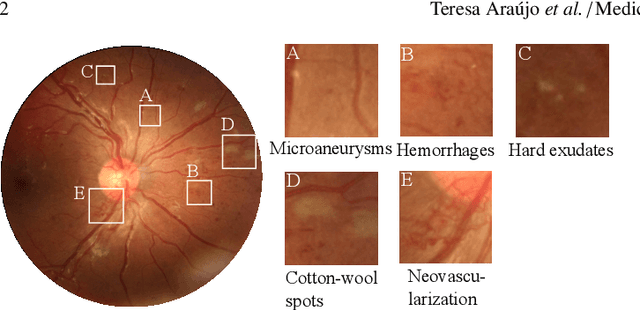
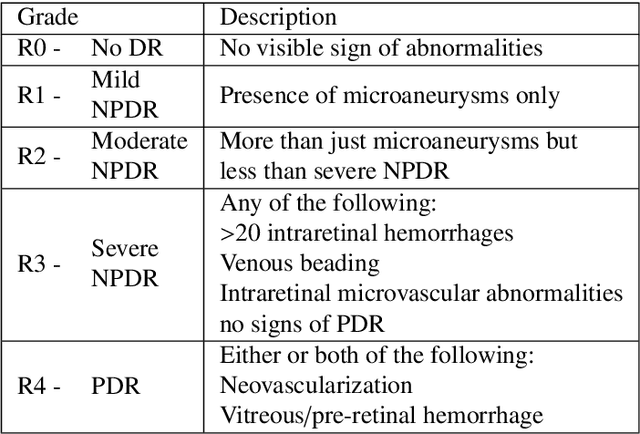
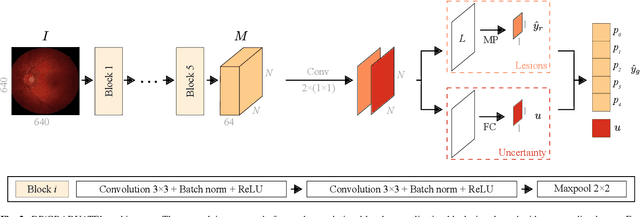
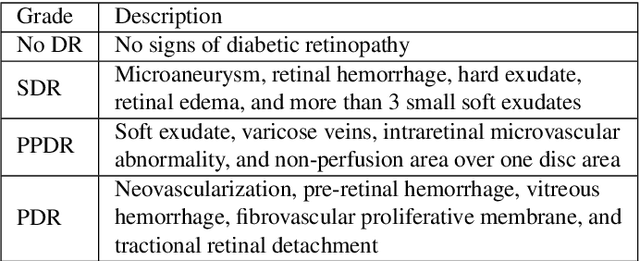
Abstract:Diabetic retinopathy (DR) grading is crucial in determining the patients' adequate treatment and follow up, but the screening process can be tiresome and prone to errors. Deep learning approaches have shown promising performance as computer-aided diagnosis(CAD) systems, but their black-box behaviour hinders the clinical application. We propose DR$\vert$GRADUATE, a novel deep learning-based DR grading CAD system that supports its decision by providing a medically interpretable explanation and an estimation of how uncertain that prediction is, allowing the ophthalmologist to measure how much that decision should be trusted. We designed DR$\vert$GRADUATE taking into account the ordinal nature of the DR grading problem. A novel Gaussian-sampling approach built upon a Multiple Instance Learning framework allow DR$\vert$GRADUATE to infer an image grade associated with an explanation map and a prediction uncertainty while being trained only with image-wise labels. DR$\vert$GRADUATE was trained on the Kaggle training set and evaluated across multiple datasets. In DR grading, a quadratic-weighted Cohen's kappa (QWK) between 0.71 and 0.84 was achieved in five different datasets. We show that high QWK values occur for images with low prediction uncertainty, thus indicating that this uncertainty is a valid measure of the predictions' quality. Further, bad quality images are generally associated with higher uncertainties, showing that images not suitable for diagnosis indeed lead to less trustworthy predictions. Additionally, tests on unfamiliar medical image data types suggest that DR$\vert$GRADUATE allows outlier detection. The attention maps generally highlight regions of interest for diagnosis. These results show the great potential of DR$\vert$GRADUATE as a second-opinion system in DR severity grading.
Did you miss it? Automatic lung nodule detection combined with gaze information improves radiologists' screening performance
Oct 09, 2019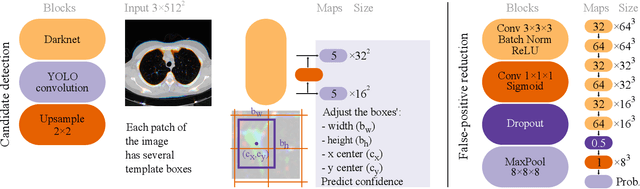
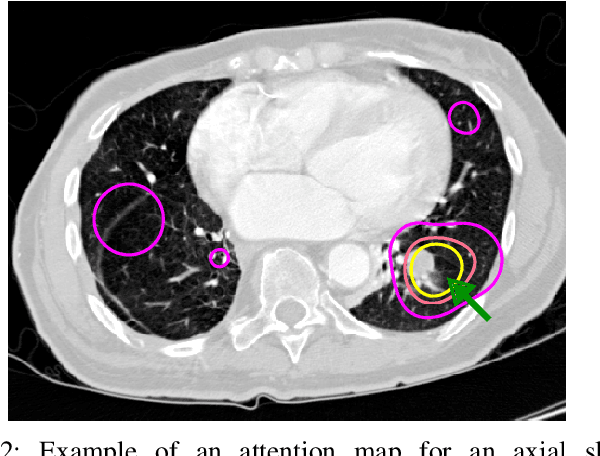
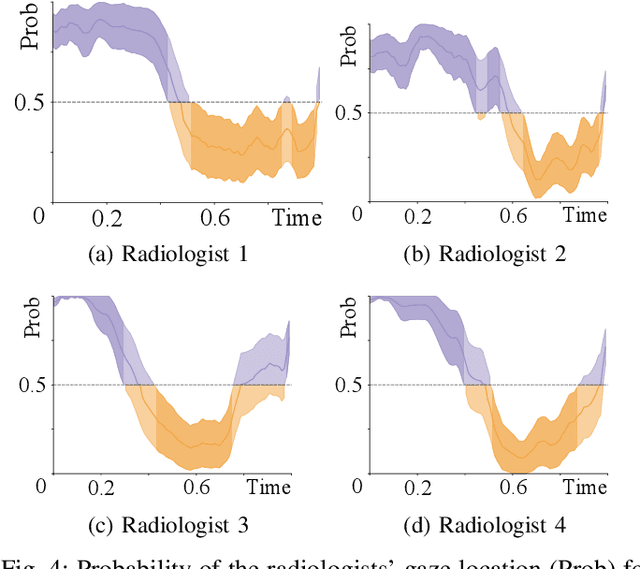
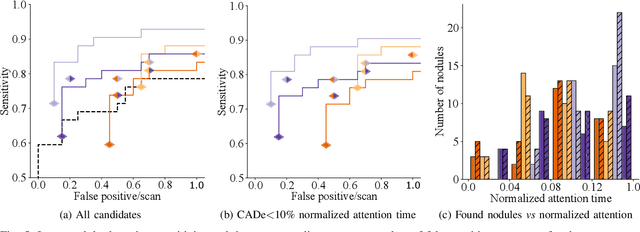
Abstract:Early diagnosis of lung cancer via computed tomography can significantly reduce the morbidity and mortality rates associated with the pathology. However, search lung nodules is a high complexity task, which affects the success of screening programs. Whilst computer-aided detection systems can be used as second observers, they may bias radiologists and introduce significant time overheads. With this in mind, this study assesses the potential of using gaze information for integrating automatic detection systems in the clinical practice. For that purpose, 4 radiologists were asked to annotate 20 scans from a public dataset while being monitored by an eye tracker device and an automatic lung nodule detection system was developed. Our results show that radiologists follow a similar search routine and tend to have lower fixation periods in regions where finding errors occur. The overall detection sensitivity of the specialists was 0.67$\pm$0.07, whereas the system achieved 0.69. Combining the annotations of one radiologist with the automatic system significantly improves the detection performance to similar levels of two annotators. Likewise, combining the findings of radiologist with the detection algorithm only for low fixation regions still significantly improves the detection sensitivity without increasing the number of false-positives. The combination of the automatic system with the gaze information allows to mitigate possible errors of the radiologist without some of the issues usually associated with automatic detection system.
O-MedAL: Online Active Deep Learning for Medical Image Analysis
Aug 28, 2019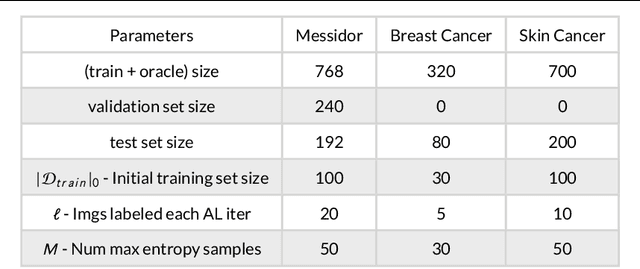
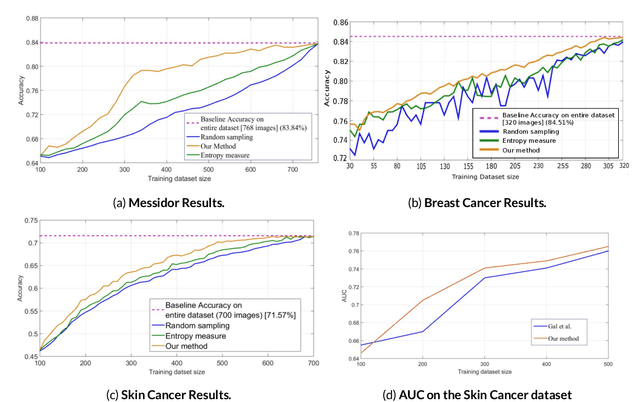
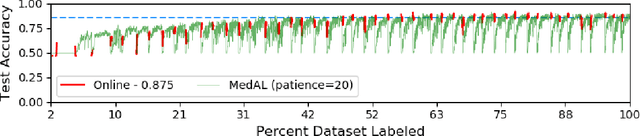
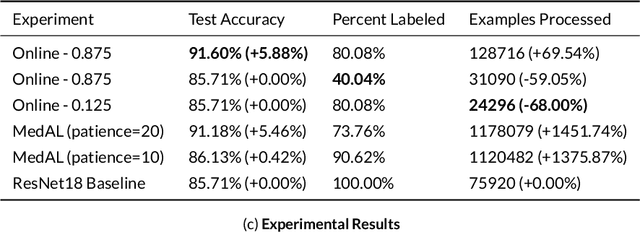
Abstract:Active Learning methods create an optimized and labeled training set from unlabeled data. We introduce a novel Online Active Deep Learning method for Medical Image Analysis. We extend our MedAL active learning framework to present new results in this paper. Experiments on three medical image datasets show that our novel online active learning model requires significantly less labelings, is more accurate, and is more robust to class imbalances than existing methods. Our method is also more accurate and computationally efficient than the baseline model. Compared to random sampling and uncertainty sampling, the method uses 275 and 200 (out of 768) fewer labeled examples, respectively. For Diabetic Retinopathy detection, our method attains a 5.88% accuracy improvement over the baseline model when 80% of the dataset is labeled, and the model reaches baseline accuracy when only 40% is labeled.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge