Mitko Veta
Tissue Cross-Section and Pen Marking Segmentation in Whole Slide Images
Jan 24, 2024Abstract:Tissue segmentation is a routine preprocessing step to reduce the computational cost of whole slide image (WSI) analysis by excluding background regions. Traditional image processing techniques are commonly used for tissue segmentation, but often require manual adjustments to parameter values for atypical cases, fail to exclude all slide and scanning artifacts from the background, and are unable to segment adipose tissue. Pen marking artifacts in particular can be a potential source of bias for subsequent analyses if not removed. In addition, several applications require the separation of individual cross-sections, which can be challenging due to tissue fragmentation and adjacent positioning. To address these problems, we develop a convolutional neural network for tissue and pen marking segmentation using a dataset of 200 H&E stained WSIs. For separating tissue cross-sections, we propose a novel post-processing method based on clustering predicted centroid locations of the cross-sections in a 2D histogram. On an independent test set, the model achieved a mean Dice score of 0.981$\pm$0.033 for tissue segmentation and a mean Dice score of 0.912$\pm$0.090 for pen marking segmentation. The mean absolute difference between the number of annotated and separated cross-sections was 0.075$\pm$0.350. Our results demonstrate that the proposed model can accurately segment H&E stained tissue cross-sections and pen markings in WSIs while being robust to many common slide and scanning artifacts. The model with trained model parameters and post-processing method are made publicly available as a Python package called SlideSegmenter.
Domain generalization across tumor types, laboratories, and species -- insights from the 2022 edition of the Mitosis Domain Generalization Challenge
Sep 27, 2023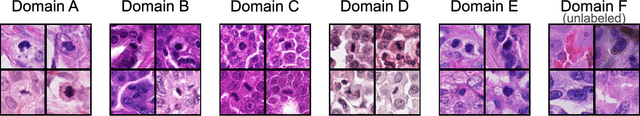
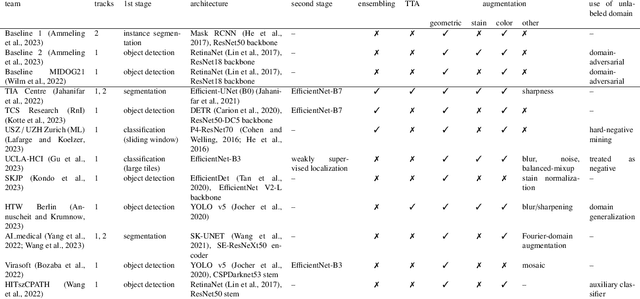
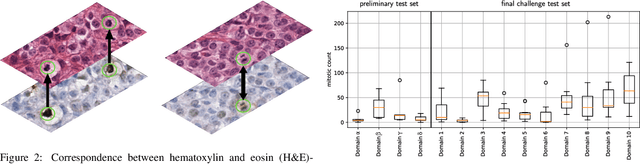
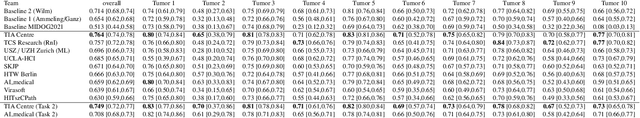
Abstract:Recognition of mitotic figures in histologic tumor specimens is highly relevant to patient outcome assessment. This task is challenging for algorithms and human experts alike, with deterioration of algorithmic performance under shifts in image representations. Considerable covariate shifts occur when assessment is performed on different tumor types, images are acquired using different digitization devices, or specimens are produced in different laboratories. This observation motivated the inception of the 2022 challenge on MItosis Domain Generalization (MIDOG 2022). The challenge provided annotated histologic tumor images from six different domains and evaluated the algorithmic approaches for mitotic figure detection provided by nine challenge participants on ten independent domains. Ground truth for mitotic figure detection was established in two ways: a three-expert consensus and an independent, immunohistochemistry-assisted set of labels. This work represents an overview of the challenge tasks, the algorithmic strategies employed by the participants, and potential factors contributing to their success. With an $F_1$ score of 0.764 for the top-performing team, we summarize that domain generalization across various tumor domains is possible with today's deep learning-based recognition pipelines. When assessed against the immunohistochemistry-assisted reference standard, all methods resulted in reduced recall scores, but with only minor changes in the order of participants in the ranking.
A Deep Learning Approach Utilizing Covariance Matrix Analysis for the ISBI Edited MRS Reconstruction Challenge
Jun 05, 2023Abstract:This work proposes a method to accelerate the acquisition of high-quality edited magnetic resonance spectroscopy (MRS) scans using machine learning models taking the sample covariance matrix as input. The method is invariant to the number of transients and robust to noisy input data for both synthetic as well as in-vivo scenarios.
Deep Learning for Detection and Localization of B-Lines in Lung Ultrasound
Feb 15, 2023



Abstract:Lung ultrasound (LUS) is an important imaging modality used by emergency physicians to assess pulmonary congestion at the patient bedside. B-line artifacts in LUS videos are key findings associated with pulmonary congestion. Not only can the interpretation of LUS be challenging for novice operators, but visual quantification of B-lines remains subject to observer variability. In this work, we investigate the strengths and weaknesses of multiple deep learning approaches for automated B-line detection and localization in LUS videos. We curate and publish, BEDLUS, a new ultrasound dataset comprising 1,419 videos from 113 patients with a total of 15,755 expert-annotated B-lines. Based on this dataset, we present a benchmark of established deep learning methods applied to the task of B-line detection. To pave the way for interpretable quantification of B-lines, we propose a novel "single-point" approach to B-line localization using only the point of origin. Our results show that (a) the area under the receiver operating characteristic curve ranges from 0.864 to 0.955 for the benchmarked detection methods, (b) within this range, the best performance is achieved by models that leverage multiple successive frames as input, and (c) the proposed single-point approach for B-line localization reaches an F1-score of 0.65, performing on par with the inter-observer agreement. The dataset and developed methods can facilitate further biomedical research on automated interpretation of lung ultrasound with the potential to expand the clinical utility.
LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset
Jan 16, 2023



Abstract:We introduce LYSTO, the Lymphocyte Assessment Hackathon, which was held in conjunction with the MICCAI 2019 Conference in Shenzen (China). The competition required participants to automatically assess the number of lymphocytes, in particular T-cells, in histopathological images of colon, breast, and prostate cancer stained with CD3 and CD8 immunohistochemistry. Differently from other challenges setup in medical image analysis, LYSTO participants were solely given a few hours to address this problem. In this paper, we describe the goal and the multi-phase organization of the hackathon; we describe the proposed methods and the on-site results. Additionally, we present post-competition results where we show how the presented methods perform on an independent set of lung cancer slides, which was not part of the initial competition, as well as a comparison on lymphocyte assessment between presented methods and a panel of pathologists. We show that some of the participants were capable to achieve pathologist-level performance at lymphocyte assessment. After the hackathon, LYSTO was left as a lightweight plug-and-play benchmark dataset on grand-challenge website, together with an automatic evaluation platform. LYSTO has supported a number of research in lymphocyte assessment in oncology. LYSTO will be a long-lasting educational challenge for deep learning and digital pathology, it is available at https://lysto.grand-challenge.org/.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022
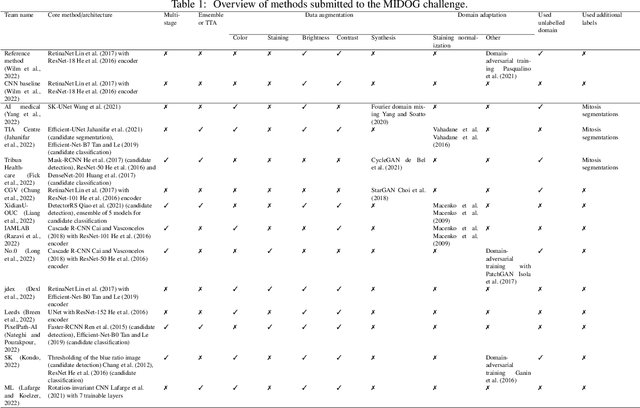
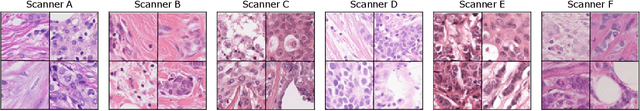
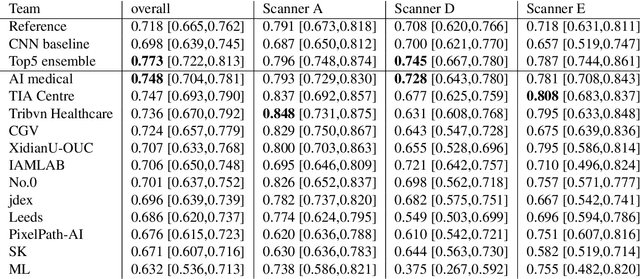
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
Optimized Automated Cardiac MR Scar Quantification with GAN-Based Data Augmentation
Sep 27, 2021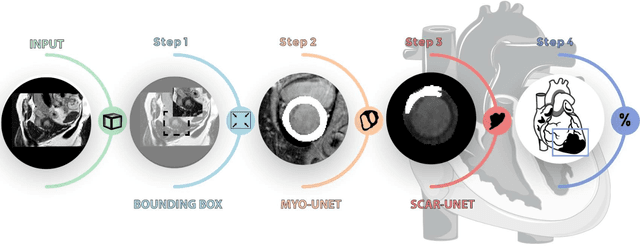
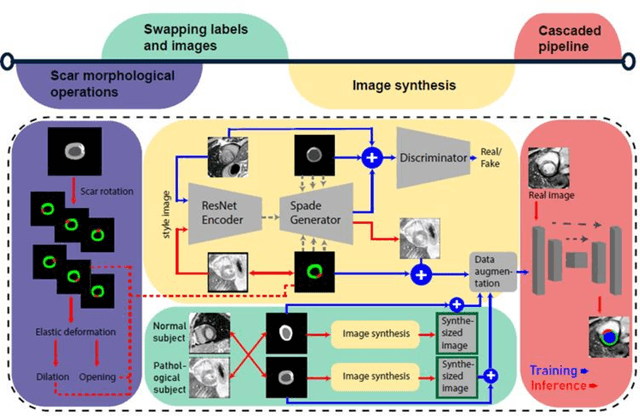
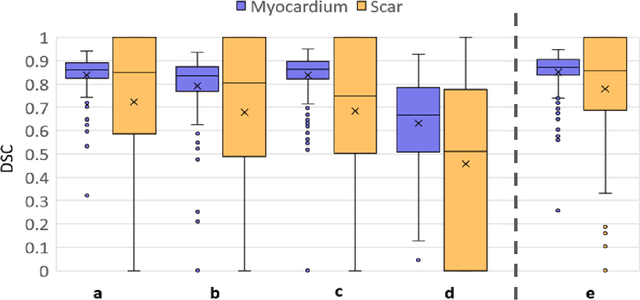
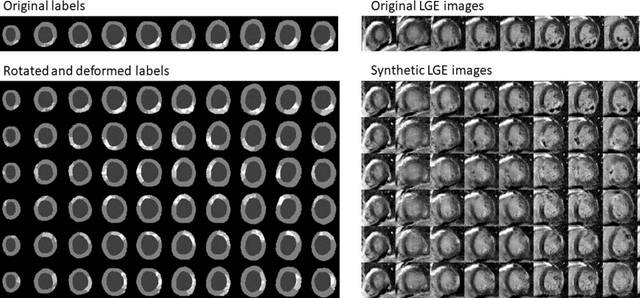
Abstract:Background: The clinical utility of late gadolinium enhancement (LGE) cardiac MRI is limited by the lack of standardization, and time-consuming postprocessing. In this work, we tested the hypothesis that a cascaded deep learning pipeline trained with augmentation by synthetically generated data would improve model accuracy and robustness for automated scar quantification. Methods: A cascaded pipeline consisting of three consecutive neural networks is proposed, starting with a bounding box regression network to identify a region of interest around the left ventricular (LV) myocardium. Two further nnU-Net models are then used to segment the myocardium and, if present, scar. The models were trained on the data from the EMIDEC challenge, supplemented with an extensive synthetic dataset generated with a conditional GAN. Results: The cascaded pipeline significantly outperformed a single nnU-Net directly segmenting both the myocardium (mean Dice similarity coefficient (DSC) (standard deviation (SD)): 0.84 (0.09) vs 0.63 (0.20), p < 0.01) and scar (DSC: 0.72 (0.34) vs 0.46 (0.39), p < 0.01) on a per-slice level. The inclusion of the synthetic data as data augmentation during training improved the scar segmentation DSC by 0.06 (p < 0.01). The mean DSC per-subject on the challenge test set, for the cascaded pipeline augmented by synthetic generated data, was 0.86 (0.03) and 0.67 (0.29) for myocardium and scar, respectively. Conclusion: A cascaded deep learning-based pipeline trained with augmentation by synthetically generated data leads to myocardium and scar segmentations that are similar to the manual operator, and outperforms direct segmentation without the synthetic images.
Quantifying the Scanner-Induced Domain Gap in Mitosis Detection
Mar 30, 2021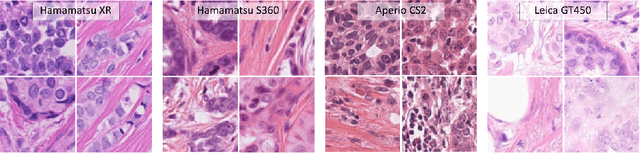
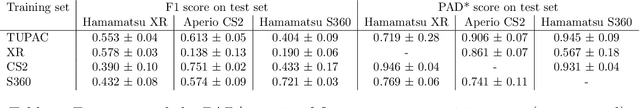
Abstract:Automated detection of mitotic figures in histopathology images has seen vast improvements, thanks to modern deep learning-based pipelines. Application of these methods, however, is in practice limited by strong variability of images between labs. This results in a domain shift of the images, which causes a performance drop of the models. Hypothesizing that the scanner device plays a decisive role in this effect, we evaluated the susceptibility of a standard mitosis detection approach to the domain shift introduced by using a different whole slide scanner. Our work is based on the MICCAI-MIDOG challenge 2021 data set, which includes 200 tumor cases of human breast cancer and four scanners. Our work indicates that the domain shift induced not by biochemical variability but purely by the choice of acquisition device is underestimated so far. Models trained on images of the same scanner yielded an average F1 score of 0.683, while models trained on a single other scanner only yielded an average F1 score of 0.325. Training on another multi-domain mitosis dataset led to mean F1 scores of 0.52. We found this not to be reflected by domain-shifts measured as proxy A distance-derived metric.
Corneal Pachymetry by AS-OCT after Descemet's Membrane Endothelial Keratoplasty
Feb 15, 2021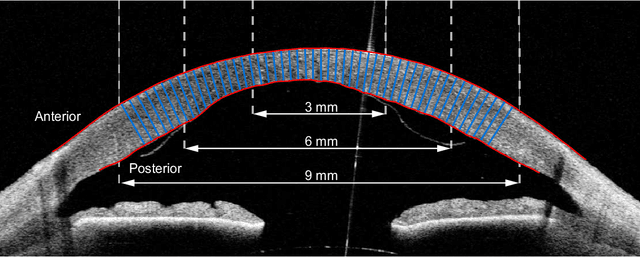
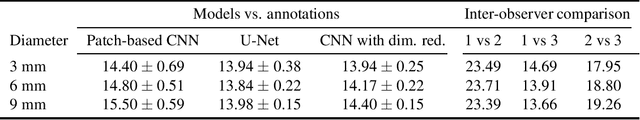
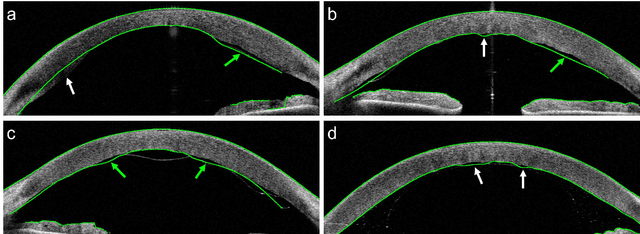
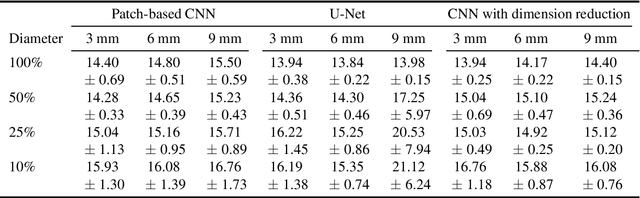
Abstract:Corneal thickness (pachymetry) maps can be used to monitor restoration of corneal endothelial function, for example after Descemet's membrane endothelial keratoplasty (DMEK). Automated delineation of the corneal interfaces in anterior segment optical coherence tomography (AS-OCT) can be challenging for corneas that are irregularly shaped due to pathology, or as a consequence of surgery, leading to incorrect thickness measurements. In this research, deep learning is used to automatically delineate the corneal interfaces and measure corneal thickness with high accuracy in post-DMEK AS-OCT B-scans. Three different deep learning strategies were developed based on 960 B-scans from 68 patients. On an independent test set of 320 B-scans, corneal thickness could be measured with an error of 13.98 to 15.50 micrometer for the central 9 mm range, which is less than 3% of the average corneal thickness. The accurate thickness measurements were used to construct detailed pachymetry maps. Moreover, follow-up scans could be registered based on anatomical landmarks to obtain differential pachymetry maps. These maps may enable a more comprehensive understanding of the restoration of the endothelial function after DMEK, where thickness often varies throughout different regions of the cornea, and subsequently contribute to a standardized postoperative regime.
Physics-informed neural networks for myocardial perfusion MRI quantification
Dec 07, 2020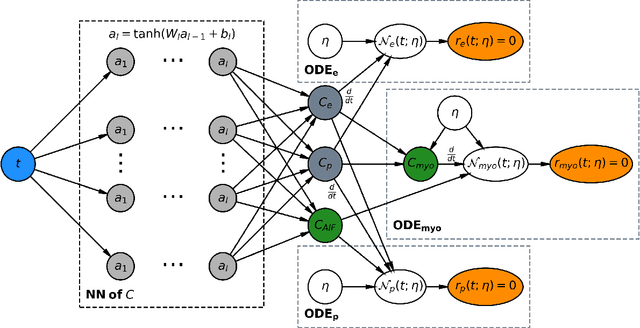
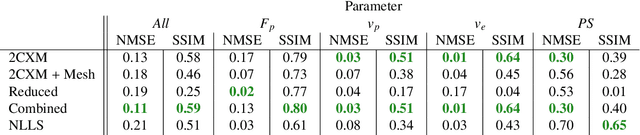
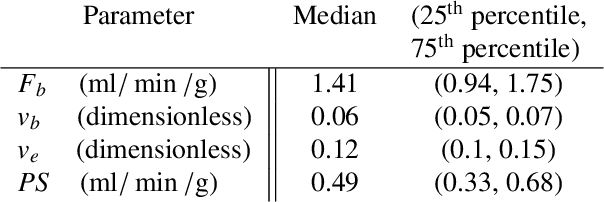
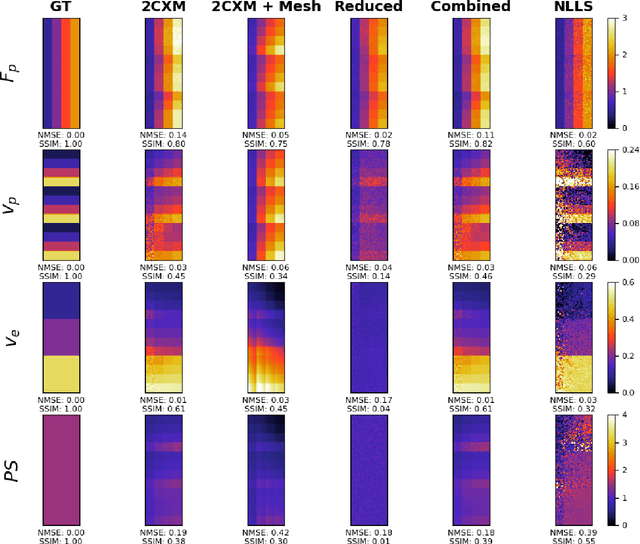
Abstract:Tracer-kinetic models allow for the quantification of kinetic parameters such as blood flow from dynamic contrast-enhanced magnetic resonance (MR) images. Fitting the observed data with multi-compartment exchange models is desirable, as they are physiologically plausible and resolve directly for blood flow and microvascular function. However, the reliability of model fitting is limited by the low signal-to-noise ratio, temporal resolution, and acquisition length. This may result in inaccurate parameter estimates. This study introduces physics-informed neural networks (PINNs) as a means to perform myocardial perfusion MR quantification, which provides a versatile scheme for the inference of kinetic parameters. These neural networks can be trained to fit the observed perfusion MR data while respecting the underlying physical conservation laws described by a multi-compartment exchange model. Here, we provide a framework for the implementation of PINNs in myocardial perfusion MR. The approach is validated both in silico and in vivo. In the in silico study, an overall reduction in mean-squared error with the ground-truth parameters was observed compared to a standard non-linear least squares fitting approach. The in vivo study demonstrates that the method produces parameter values comparable to those previously found in literature, as well as providing parameter maps which match the clinical diagnosis of patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge