Maxim A. Ziatdinov
Integrating Predictive and Generative Capabilities by Latent Space Design via the DKL-VAE Model
Mar 04, 2025Abstract:We introduce a Deep Kernel Learning Variational Autoencoder (VAE-DKL) framework that integrates the generative power of a Variational Autoencoder (VAE) with the predictive nature of Deep Kernel Learning (DKL). The VAE learns a latent representation of high-dimensional data, enabling the generation of novel structures, while DKL refines this latent space by structuring it in alignment with target properties through Gaussian Process (GP) regression. This approach preserves the generative capabilities of the VAE while enhancing its latent space for GP-based property prediction. We evaluate the framework on two datasets: a structured card dataset with predefined variational factors and the QM9 molecular dataset, where enthalpy serves as the target function for optimization. The model demonstrates high-precision property prediction and enables the generation of novel out-of-training subset structures with desired characteristics. The VAE-DKL framework offers a promising approach for high-throughput material discovery and molecular design, balancing structured latent space organization with generative flexibility.
Reward driven workflows for unsupervised explainable analysis of phases and ferroic variants from atomically resolved imaging data
Nov 19, 2024



Abstract:Rapid progress in aberration corrected electron microscopy necessitates development of robust methods for the identification of phases, ferroic variants, and other pertinent aspects of materials structure from imaging data. While unsupervised methods for clustering and classification are widely used for these tasks, their performance can be sensitive to hyperparameter selection in the analysis workflow. In this study, we explore the effects of descriptors and hyperparameters on the capability of unsupervised ML methods to distill local structural information, exemplified by discovery of polarization and lattice distortion in Sm doped BiFeO3 (BFO) thin films. We demonstrate that a reward-driven approach can be used to optimize these key hyperparameters across the full workflow, where rewards were designed to reflect domain wall continuity and straightness, ensuring that the analysis aligns with the material's physical behavior. This approach allows us to discover local descriptors that are best aligned with the specific physical behavior, providing insight into the fundamental physics of materials. We further extend the reward driven workflows to disentangle structural factors of variation via optimized variational autoencoder (VAE). Finally, the importance of well-defined rewards was explored as a quantifiable measure of success of the workflow.
Bayesian Co-navigation: Dynamic Designing of the Materials Digital Twins via Active Learning
Apr 19, 2024



Abstract:Scientific advancement is universally based on the dynamic interplay between theoretical insights, modelling, and experimental discoveries. However, this feedback loop is often slow, including delayed community interactions and the gradual integration of experimental data into theoretical frameworks. This challenge is particularly exacerbated in domains dealing with high-dimensional object spaces, such as molecules and complex microstructures. Hence, the integration of theory within automated and autonomous experimental setups, or theory in the loop automated experiment, is emerging as a crucial objective for accelerating scientific research. The critical aspect is not only to use theory but also on-the-fly theory updates during the experiment. Here, we introduce a method for integrating theory into the loop through Bayesian co-navigation of theoretical model space and experimentation. Our approach leverages the concurrent development of surrogate models for both simulation and experimental domains at the rates determined by latencies and costs of experiments and computation, alongside the adjustment of control parameters within theoretical models to minimize epistemic uncertainty over the experimental object spaces. This methodology facilitates the creation of digital twins of material structures, encompassing both the surrogate model of behavior that includes the correlative part and the theoretical model itself. While demonstrated here within the context of functional responses in ferroelectric materials, our approach holds promise for broader applications, the exploration of optical properties in nanoclusters, microstructure-dependent properties in complex materials, and properties of molecular systems. The analysis code that supports the funding is publicly available at https://github.com/Slautin/2024_Co-navigation/tree/main
Multimodal Co-orchestration for Exploring Structure-Property Relationships in Combinatorial Libraries via Multi-Task Bayesian Optimization
Feb 03, 2024Abstract:The rapid growth of automated and autonomous instrumentations brings forth an opportunity for the co-orchestration of multimodal tools, equipped with multiple sequential detection methods, or several characterization tools to explore identical samples. This can be exemplified by the combinatorial libraries that can be explored in multiple locations by multiple tools simultaneously, or downstream characterization in automated synthesis systems. In the co-orchestration approaches, information gained in one modality should accelerate the discovery of other modalities. Correspondingly, the orchestrating agent should select the measurement modality based on the anticipated knowledge gain and measurement cost. Here, we propose and implement a co-orchestration approach for conducting measurements with complex observables such as spectra or images. The method relies on combining dimensionality reduction by variational autoencoders with representation learning for control over the latent space structure, and integrated into iterative workflow via multi-task Gaussian Processes (GP). This approach further allows for the native incorporation of the system's physics via a probabilistic model as a mean function of the GP. We illustrated this method for different modalities of piezoresponse force microscopy and micro-Raman on combinatorial $Sm-BiFeO_3$ library. However, the proposed framework is general and can be extended to multiple measurement modalities and arbitrary dimensionality of measured signals. The analysis code that supports the funding is publicly available at https://github.com/Slautin/2024_Co-orchestration.
A dynamic Bayesian optimized active recommender system for curiosity-driven Human-in-the-loop automated experiments
Apr 05, 2023Abstract:Optimization of experimental materials synthesis and characterization through active learning methods has been growing over the last decade, with examples ranging from measurements of diffraction on combinatorial alloys at synchrotrons, to searches through chemical space with automated synthesis robots for perovskites. In virtually all cases, the target property of interest for optimization is defined apriori with limited human feedback during operation. In contrast, here we present the development of a new type of human in the loop experimental workflow, via a Bayesian optimized active recommender system (BOARS), to shape targets on the fly, employing human feedback. We showcase examples of this framework applied to pre-acquired piezoresponse force spectroscopy of a ferroelectric thin film, and then implement this in real time on an atomic force microscope, where the optimization proceeds to find symmetric piezoresponse amplitude hysteresis loops. It is found that such features appear more affected by subsurface defects than the local domain structure. This work shows the utility of human-augmented machine learning approaches for curiosity-driven exploration of systems across experimental domains. The analysis reported here is summarized in Colab Notebook for the purpose of tutorial and application to other data: https://github.com/arpanbiswas52/varTBO
Deep Learning for Automated Experimentation in Scanning Transmission Electron Microscopy
Apr 04, 2023



Abstract:Machine learning (ML) has become critical for post-acquisition data analysis in (scanning) transmission electron microscopy, (S)TEM, imaging and spectroscopy. An emerging trend is the transition to real-time analysis and closed-loop microscope operation. The effective use of ML in electron microscopy now requires the development of strategies for microscopy-centered experiment workflow design and optimization. Here, we discuss the associated challenges with the transition to active ML, including sequential data analysis and out-of-distribution drift effects, the requirements for the edge operation, local and cloud data storage, and theory in the loop operations. Specifically, we discuss the relative contributions of human scientists and ML agents in the ideation, orchestration, and execution of experimental workflows and the need to develop universal hyper languages that can apply across multiple platforms. These considerations will collectively inform the operationalization of ML in next-generation experimentation.
Deep Kernel Methods Learn Better: From Cards to Process Optimization
Mar 25, 2023Abstract:The ability of deep learning methods to perform classification and regression tasks relies heavily on their capacity to uncover manifolds in high-dimensional data spaces and project them into low-dimensional representation spaces. In this study, we investigate the structure and character of the manifolds generated by classical variational autoencoder (VAE) approaches and deep kernel learning (DKL). In the former case, the structure of the latent space is determined by the properties of the input data alone, while in the latter, the latent manifold forms as a result of an active learning process that balances the data distribution and target functionalities. We show that DKL with active learning can produce a more compact and smooth latent space which is more conducive to optimization compared to previously reported methods, such as the VAE. We demonstrate this behavior using a simple cards data set and extend it to the optimization of domain-generated trajectories in physical systems. Our findings suggest that latent manifolds constructed through active learning have a more beneficial structure for optimization problems, especially in feature-rich target-poor scenarios that are common in domain sciences, such as materials synthesis, energy storage, and molecular discovery. The jupyter notebooks that encapsulate the complete analysis accompany the article.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Discovery of structure-property relations for molecules via hypothesis-driven active learning over the chemical space
Jan 06, 2023



Abstract:Discovery of the molecular candidates for applications in drug targets, biomolecular systems, catalysts, photovoltaics, organic electronics, and batteries, necessitates development of machine learning algorithms capable of rapid exploration of the chemical spaces targeting the desired functionalities. Here we introduce a novel approach for the active learning over the chemical spaces based on hypothesis learning. We construct the hypotheses on the possible relationships between structures and functionalities of interest based on a small subset of data and introduce them as (probabilistic) mean functions for the Gaussian process. This approach combines the elements from the symbolic regression methods such as SISSO and active learning into a single framework. Here, we demonstrate it for the QM9 dataset, but it can be applied more broadly to datasets from both domains of molecular and solid-state materials sciences.
Automated and Autonomous Experiment in Electron and Scanning Probe Microscopy
Mar 22, 2021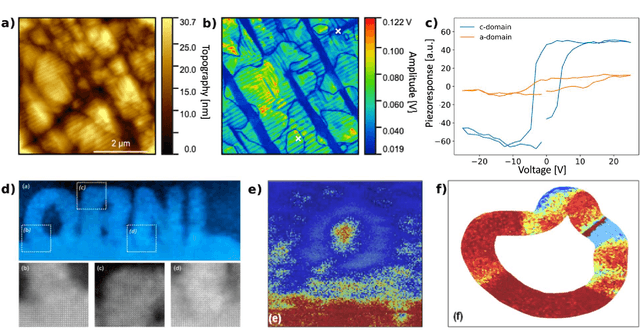
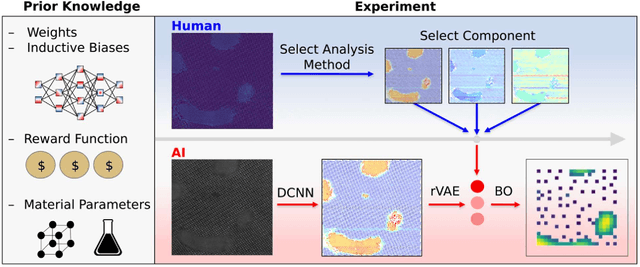
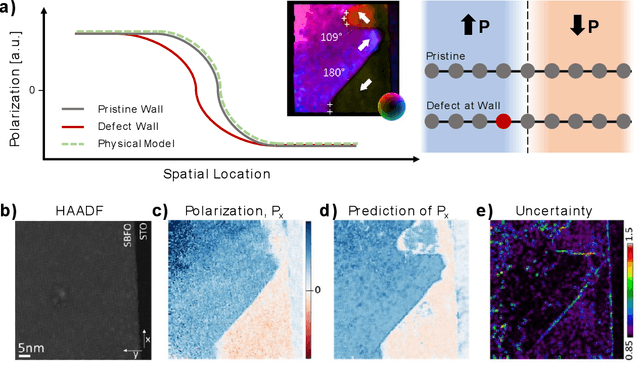
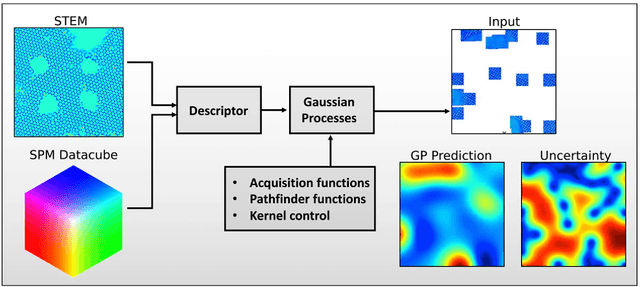
Abstract:Machine learning and artificial intelligence (ML/AI) are rapidly becoming an indispensable part of physics research, with domain applications ranging from theory and materials prediction to high-throughput data analysis. In parallel, the recent successes in applying ML/AI methods for autonomous systems from robotics through self-driving cars to organic and inorganic synthesis are generating enthusiasm for the potential of these techniques to enable automated and autonomous experiment (AE) in imaging. Here, we aim to analyze the major pathways towards AE in imaging methods with sequential image formation mechanisms, focusing on scanning probe microscopy (SPM) and (scanning) transmission electron microscopy ((S)TEM). We argue that automated experiments should necessarily be discussed in a broader context of the general domain knowledge that both informs the experiment and is increased as the result of the experiment. As such, this analysis should explore the human and ML/AI roles prior to and during the experiment, and consider the latencies, biases, and knowledge priors of the decision-making process. Similarly, such discussion should include the limitations of the existing imaging systems, including intrinsic latencies, non-idealities and drifts comprising both correctable and stochastic components. We further pose that the role of the AE in microscopy is not the exclusion of human operators (as is the case for autonomous driving), but rather automation of routine operations such as microscope tuning, etc., prior to the experiment, and conversion of low latency decision making processes on the time scale spanning from image acquisition to human-level high-order experiment planning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge