Ivo F. Sbalzarini
Push-Forward Signed Distance Functions enable interpretable and robust continuous shape quantification
Oct 28, 2024Abstract:We introduce the Push-Forward Signed Distance Morphometric (PF-SDM), a novel method for shape quantification in biomedical imaging that is continuous, interpretable, and invariant to shape-preserving transformations. PF-SDM effectively captures the geometric properties of shapes, including their topological skeletons and radial symmetries. This results in a robust and interpretable shape descriptor that generalizes to capture temporal shape dynamics. Importantly, PF-SDM avoids certain issues of previous geometric morphometrics, like Elliptical Fourier Analysis and Generalized Procrustes Analysis, such as coefficient correlations and landmark choices. We present the PF-SDM theory, provide a practically computable algorithm, and benchmark it on synthetic data.
Learning locally dominant force balances in active particle systems
Jul 27, 2023Abstract:We use a combination of unsupervised clustering and sparsity-promoting inference algorithms to learn locally dominant force balances that explain macroscopic pattern formation in self-organized active particle systems. The self-organized emergence of macroscopic patterns from microscopic interactions between self-propelled particles can be widely observed nature. Although hydrodynamic theories help us better understand the physical basis of this phenomenon, identifying a sufficient set of local interactions that shape, regulate, and sustain self-organized structures in active particle systems remains challenging. We investigate a classic hydrodynamic model of self-propelled particles that produces a wide variety of patterns, like asters and moving density bands. Our data-driven analysis shows that propagating bands are formed by local alignment interactions driven by density gradients, while steady-state asters are shaped by a mechanism of splay-induced negative compressibility arising from strong particle interactions. Our method also reveals analogous physical principles of pattern formation in a system where the speed of the particle is influenced by local density. This demonstrates the ability of our method to reveal physical commonalities across models. The physical mechanisms inferred from the data are in excellent agreement with analytical scaling arguments and experimental observations.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Learning deterministic hydrodynamic equations from stochastic active particle dynamics
Jan 21, 2022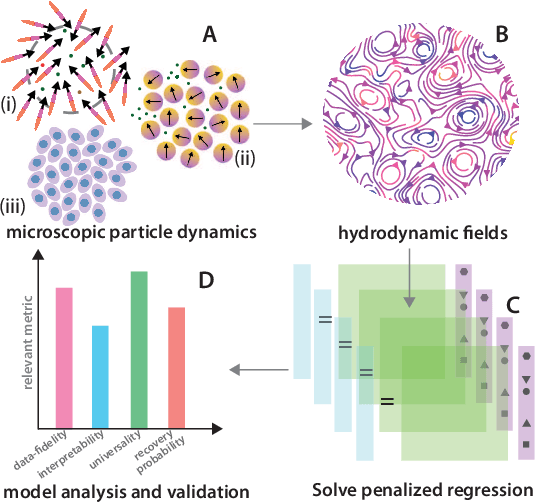
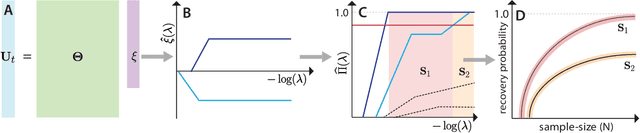
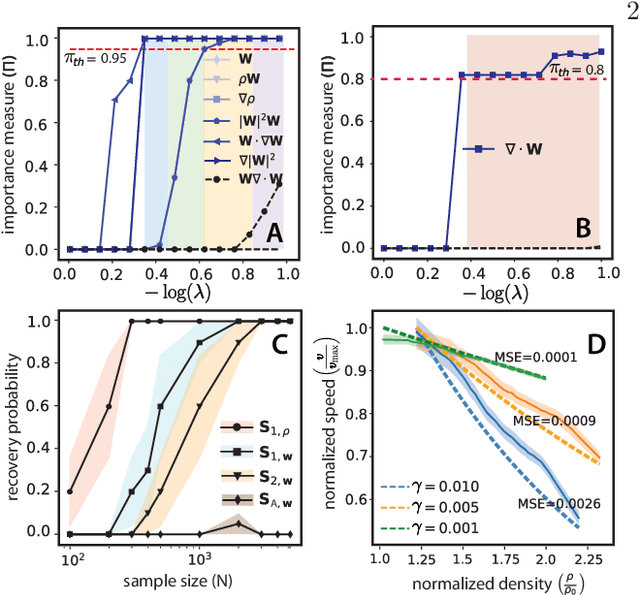
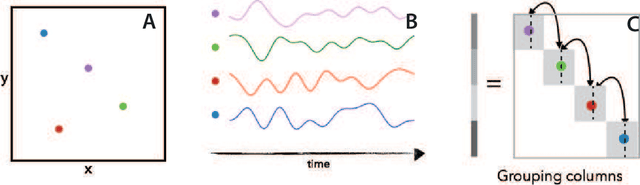
Abstract:We present a principled data-driven strategy for learning deterministic hydrodynamic models directly from stochastic non-equilibrium active particle trajectories. We apply our method to learning a hydrodynamic model for the propagating density lanes observed in self-propelled particle systems and to learning a continuum description of cell dynamics in epithelial tissues. We also infer from stochastic particle trajectories the latent phoretic fields driving chemotaxis. This demonstrates that statistical learning theory combined with physical priors can enable discovery of multi-scale models of non-equilibrium stochastic processes characteristic of collective movement in living systems.
Parallel Discrete Convolutions on Adaptive Particle Representations of Images
Dec 07, 2021



Abstract:We present data structures and algorithms for native implementations of discrete convolution operators over Adaptive Particle Representations (APR) of images on parallel computer architectures. The APR is a content-adaptive image representation that locally adapts the sampling resolution to the image signal. It has been developed as an alternative to pixel representations for large, sparse images as they typically occur in fluorescence microscopy. It has been shown to reduce the memory and runtime costs of storing, visualizing, and processing such images. This, however, requires that image processing natively operates on APRs, without intermediately reverting to pixels. Designing efficient and scalable APR-native image processing primitives, however, is complicated by the APR's irregular memory structure. Here, we provide the algorithmic building blocks required to efficiently and natively process APR images using a wide range of algorithms that can be formulated in terms of discrete convolutions. We show that APR convolution naturally leads to scale-adaptive algorithms that efficiently parallelize on multi-core CPU and GPU architectures. We quantify the speedups in comparison to pixel-based algorithms and convolutions on evenly sampled data. We achieve pixel-equivalent throughputs of up to 1 TB/s on a single Nvidia GeForce RTX 2080 gaming GPU, requiring up to two orders of magnitude less memory than a pixel-based implementation.
Inverse-Dirichlet Weighting Enables Reliable Training of Physics Informed Neural Networks
Jul 02, 2021



Abstract:We characterize and remedy a failure mode that may arise from multi-scale dynamics with scale imbalances during training of deep neural networks, such as Physics Informed Neural Networks (PINNs). PINNs are popular machine-learning templates that allow for seamless integration of physical equation models with data. Their training amounts to solving an optimization problem over a weighted sum of data-fidelity and equation-fidelity objectives. Conflicts between objectives can arise from scale imbalances, heteroscedasticity in the data, stiffness of the physical equation, or from catastrophic interference during sequential training. We explain the training pathology arising from this and propose a simple yet effective inverse-Dirichlet weighting strategy to alleviate the issue. We compare with Sobolev training of neural networks, providing the baseline of analytically $\boldsymbol{\epsilon}$-optimal training. We demonstrate the effectiveness of inverse-Dirichlet weighting in various applications, including a multi-scale model of active turbulence, where we show orders of magnitude improvement in accuracy and convergence over conventional PINN training. For inverse modeling using sequential training, we find that inverse-Dirichlet weighting protects a PINN against catastrophic forgetting.
STENCIL-NET: Data-driven solution-adaptive discretization of partial differential equations
Jan 18, 2021



Abstract:Numerical methods for approximately solving partial differential equations (PDE) are at the core of scientific computing. Often, this requires high-resolution or adaptive discretization grids to capture relevant spatio-temporal features in the PDE solution, e.g., in applications like turbulence, combustion, and shock propagation. Numerical approximation also requires knowing the PDE in order to construct problem-specific discretizations. Systematically deriving such solution-adaptive discrete operators, however, is a current challenge. Here we present STENCIL-NET, an artificial neural network architecture for data-driven learning of problem- and resolution-specific local discretizations of nonlinear PDEs. STENCIL-NET achieves numerically stable discretization of the operators in an unknown nonlinear PDE by spatially and temporally adaptive parametric pooling on regular Cartesian grids, and by incorporating knowledge about discrete time integration. Knowing the actual PDE is not necessary, as solution data is sufficient to train the network to learn the discrete operators. A once-trained STENCIL-NET model can be used to predict solutions of the PDE on larger spatial domains and for longer times than it was trained for, hence addressing the problem of PDE-constrained extrapolation from data. To support this claim, we present numerical experiments on long-term forecasting of chaotic PDE solutions on coarse spatio-temporal grids. We also quantify the speed-up achieved by substituting base-line numerical methods with equation-free STENCIL-NET predictions on coarser grids with little compromise on accuracy.
Learning physically consistent mathematical models from data using group sparsity
Dec 11, 2020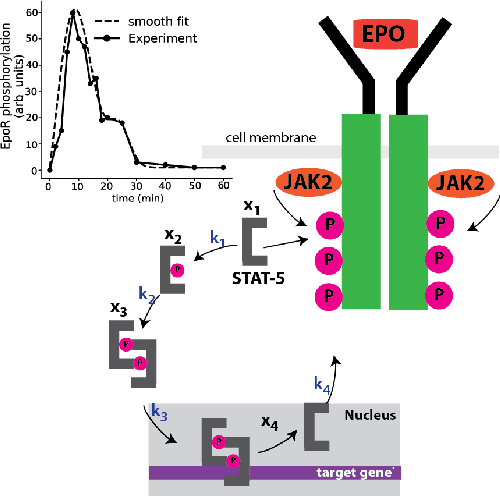
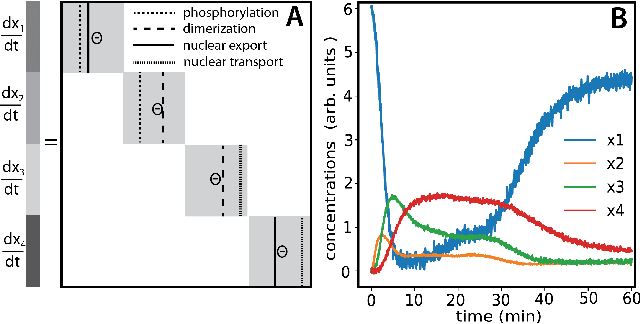
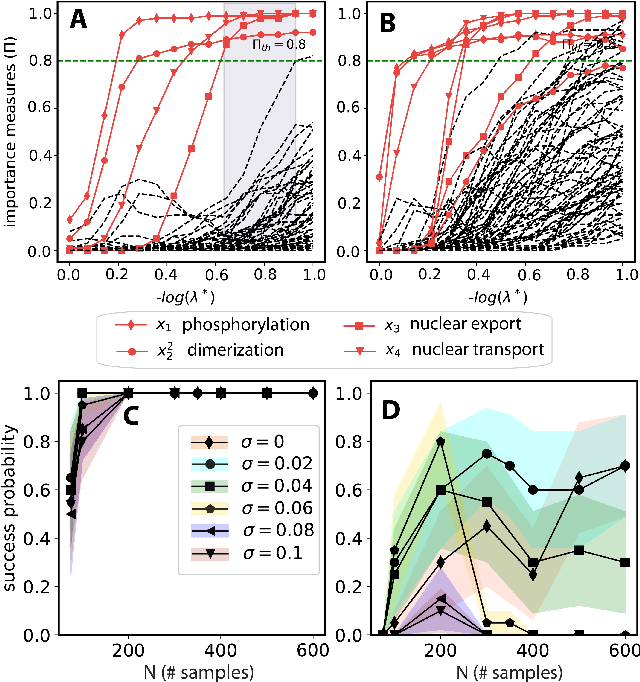
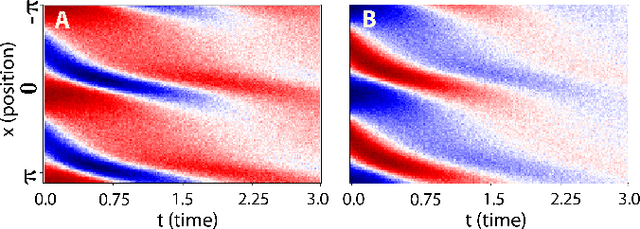
Abstract:We propose a statistical learning framework based on group-sparse regression that can be used to 1) enforce conservation laws, 2) ensure model equivalence, and 3) guarantee symmetries when learning or inferring differential-equation models from measurement data. Directly learning $\textit{interpretable}$ mathematical models from data has emerged as a valuable modeling approach. However, in areas like biology, high noise levels, sensor-induced correlations, and strong inter-system variability can render data-driven models nonsensical or physically inconsistent without additional constraints on the model structure. Hence, it is important to leverage $\textit{prior}$ knowledge from physical principles to learn "biologically plausible and physically consistent" models rather than models that simply fit the data best. We present a novel group Iterative Hard Thresholding (gIHT) algorithm and use stability selection to infer physically consistent models with minimal parameter tuning. We show several applications from systems biology that demonstrate the benefits of enforcing $\textit{priors}$ in data-driven modeling.
A robustness measure for singular point and index estimation in discretized orientation and vector fields
Sep 30, 2020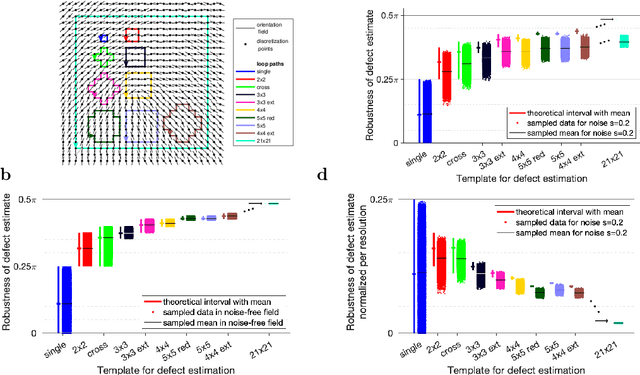
Abstract:The identification of singular points or topological defects in discretized vector fields occurs in diverse areas ranging from the polarization of the cosmic microwave background to liquid crystals to fingerprint recognition and bio-medical imaging. Due to their discrete nature, defects and their topological charge cannot depend continuously on each single vector, but they discontinuously change as soon as a vector changes by more than a threshold. Considering this threshold of admissible change at the level of vectors, we develop a robustness measure for discrete defect estimators. Here, we compare different template paths for defect estimation in discretized vector or orientation fields. Sampling prototypical vector field patterns around defects shows that the robustness increases with the length of template path, but less so in the presence of noise on the vectors. We therefore find an optimal trade-off between resolution and robustness against noise for relatively small templates, except for the "single pixel" defect analysis, which cannot exclude zero robustness. The presented robustness measure paves the way for uncertainty quantification of defects in discretized vector fields.
Stability selection enables robust learning of partial differential equations from limited noisy data
Jul 17, 2019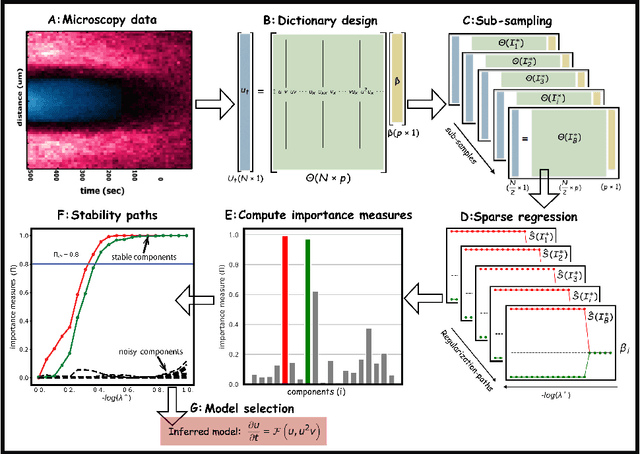
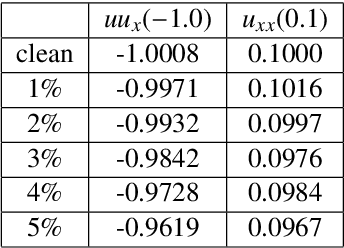
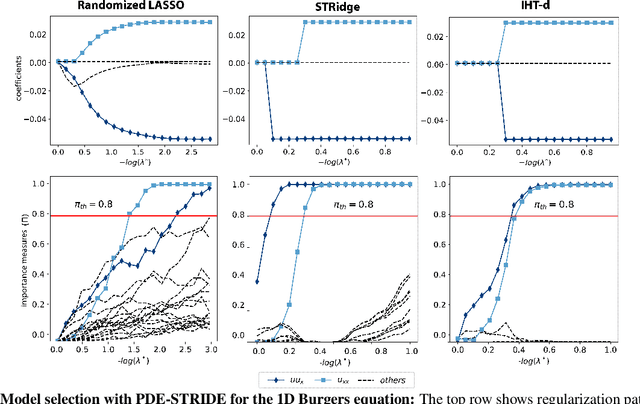
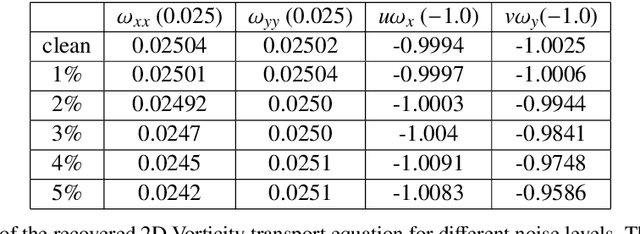
Abstract:We present a statistical learning framework for robust identification of partial differential equations from noisy spatiotemporal data. Extending previous sparse regression approaches for inferring PDE models from simulated data, we address key issues that have thus far limited the application of these methods to noisy experimental data, namely their robustness against noise and the need for manual parameter tuning. We address both points by proposing a stability-based model selection scheme to determine the level of regularization required for reproducible recovery of the underlying PDE. This avoids manual parameter tuning and provides a principled way to improve the method's robustness against noise in the data. Our stability selection approach, termed PDE-STRIDE, can be combined with any sparsity-promoting penalized regression model and provides an interpretable criterion for model component importance. We show that in particular the combination of stability selection with the iterative hard-thresholding algorithm from compressed sensing provides a fast, parameter-free, and robust computational framework for PDE inference that outperforms previous algorithmic approaches with respect to recovery accuracy, amount of data required, and robustness to noise. We illustrate the performance of our approach on a wide range of noise-corrupted simulated benchmark problems, including 1D Burgers, 2D vorticity-transport, and 3D reaction-diffusion problems. We demonstrate the practical applicability of our method on real-world data by considering a purely data-driven re-evaluation of the advective triggering hypothesis for an embryonic polarization system in C.~elegans. Using fluorescence microscopy images of C.~elegans zygotes as input data, our framework is able to recover the PDE model for the regulatory reaction-diffusion-flow network of the associated proteins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge