Sarah Akers
Revealing the Evolution of Order in Materials Microstructures Using Multi-Modal Computer Vision
Nov 15, 2024



Abstract:The development of high-performance materials for microelectronics, energy storage, and extreme environments depends on our ability to describe and direct property-defining microstructural order. Our present understanding is typically derived from laborious manual analysis of imaging and spectroscopy data, which is difficult to scale, challenging to reproduce, and lacks the ability to reveal latent associations needed for mechanistic models. Here, we demonstrate a multi-modal machine learning (ML) approach to describe order from electron microscopy analysis of the complex oxide La$_{1-x}$Sr$_x$FeO$_3$. We construct a hybrid pipeline based on fully and semi-supervised classification, allowing us to evaluate both the characteristics of each data modality and the value each modality adds to the ensemble. We observe distinct differences in the performance of uni- and multi-modal models, from which we draw general lessons in describing crystal order using computer vision.
Final Report for CHESS: Cloud, High-Performance Computing, and Edge for Science and Security
Oct 21, 2024



Abstract:Automating the theory-experiment cycle requires effective distributed workflows that utilize a computing continuum spanning lab instruments, edge sensors, computing resources at multiple facilities, data sets distributed across multiple information sources, and potentially cloud. Unfortunately, the obvious methods for constructing continuum platforms, orchestrating workflow tasks, and curating datasets over time fail to achieve scientific requirements for performance, energy, security, and reliability. Furthermore, achieving the best use of continuum resources depends upon the efficient composition and execution of workflow tasks, i.e., combinations of numerical solvers, data analytics, and machine learning. Pacific Northwest National Laboratory's LDRD "Cloud, High-Performance Computing (HPC), and Edge for Science and Security" (CHESS) has developed a set of interrelated capabilities for enabling distributed scientific workflows and curating datasets. This report describes the results and successes of CHESS from the perspective of open science.
Deep Learning for Automated Experimentation in Scanning Transmission Electron Microscopy
Apr 04, 2023



Abstract:Machine learning (ML) has become critical for post-acquisition data analysis in (scanning) transmission electron microscopy, (S)TEM, imaging and spectroscopy. An emerging trend is the transition to real-time analysis and closed-loop microscope operation. The effective use of ML in electron microscopy now requires the development of strategies for microscopy-centered experiment workflow design and optimization. Here, we discuss the associated challenges with the transition to active ML, including sequential data analysis and out-of-distribution drift effects, the requirements for the edge operation, local and cloud data storage, and theory in the loop operations. Specifically, we discuss the relative contributions of human scientists and ML agents in the ideation, orchestration, and execution of experimental workflows and the need to develop universal hyper languages that can apply across multiple platforms. These considerations will collectively inform the operationalization of ML in next-generation experimentation.
An Automated Scanning Transmission Electron Microscope Guided by Sparse Data Analytics
Sep 30, 2021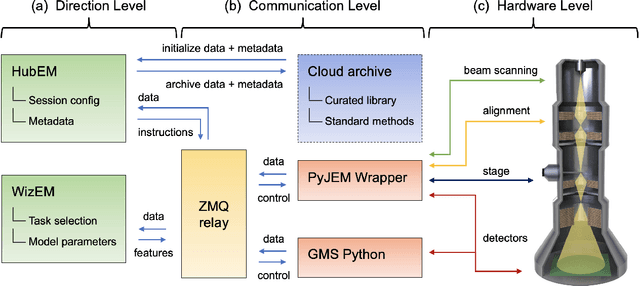
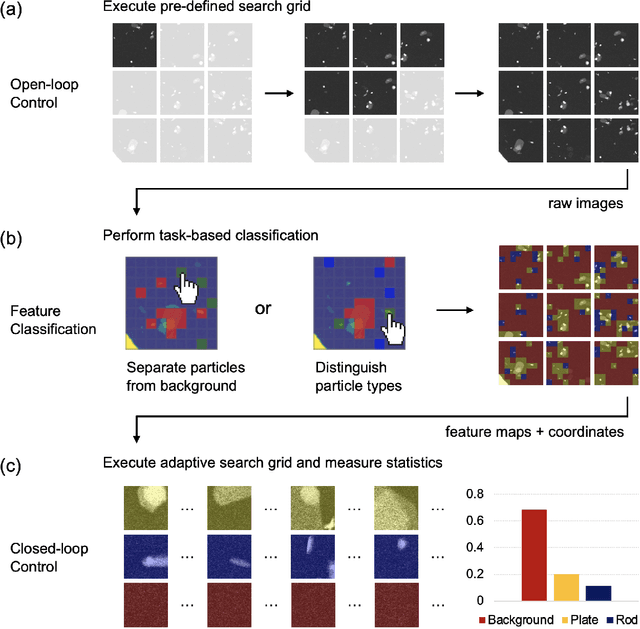
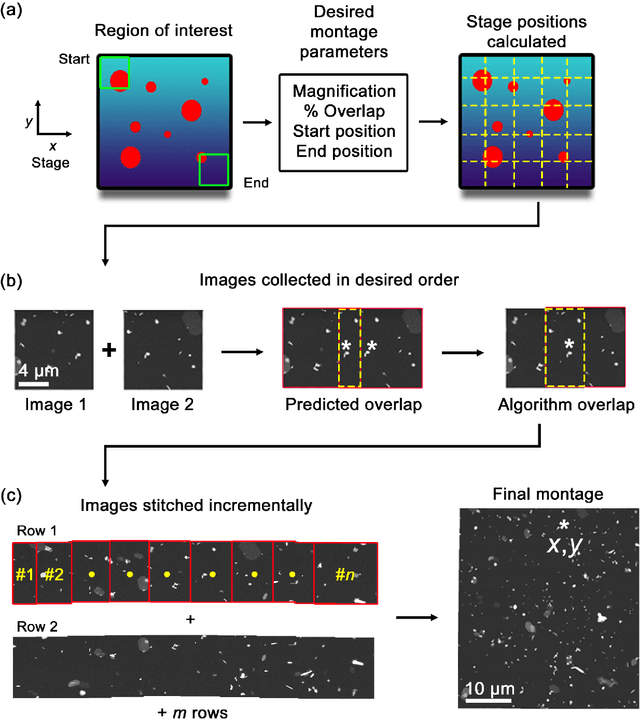
Abstract:Artificial intelligence (AI) promises to reshape scientific inquiry and enable breakthrough discoveries in areas such as energy storage, quantum computing, and biomedicine. Scanning transmission electron microscopy (STEM), a cornerstone of the study of chemical and materials systems, stands to benefit greatly from AI-driven automation. However, present barriers to low-level instrument control, as well as generalizable and interpretable feature detection, make truly automated microscopy impractical. Here, we discuss the design of a closed-loop instrument control platform guided by emerging sparse data analytics. We demonstrate how a centralized controller, informed by machine learning combining limited $a$ $priori$ knowledge and task-based discrimination, can drive on-the-fly experimental decision-making. This platform unlocks practical, automated analysis of a variety of material features, enabling new high-throughput and statistical studies.
Design of a Graphical User Interface for Few-Shot Machine Learning Classification of Electron Microscopy Data
Jul 21, 2021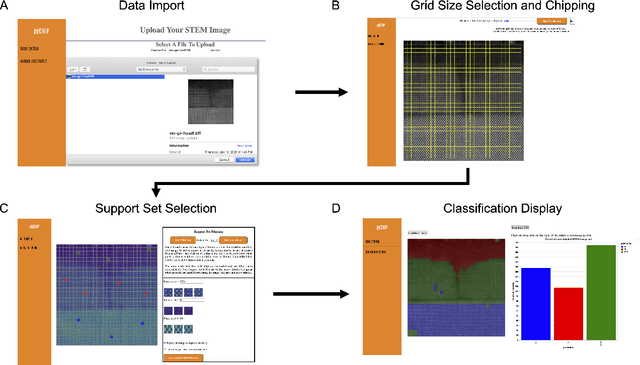
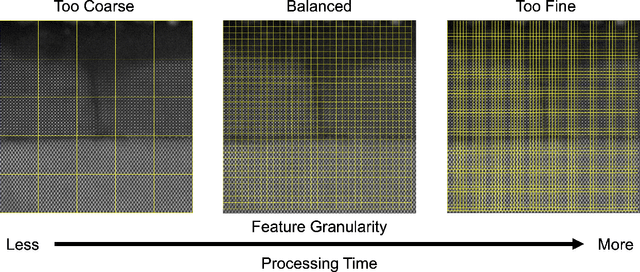

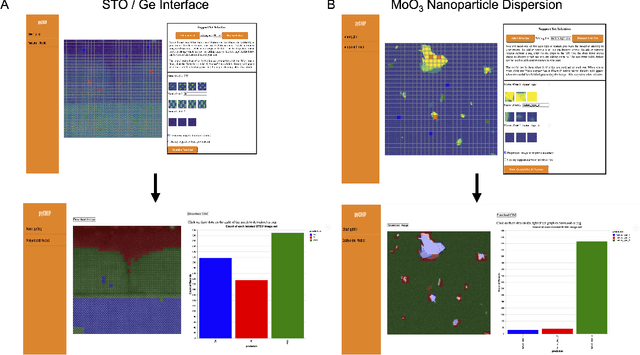
Abstract:The recent growth in data volumes produced by modern electron microscopes requires rapid, scalable, and flexible approaches to image segmentation and analysis. Few-shot machine learning, which can richly classify images from a handful of user-provided examples, is a promising route to high-throughput analysis. However, current command-line implementations of such approaches can be slow and unintuitive to use, lacking the real-time feedback necessary to perform effective classification. Here we report on the development of a Python-based graphical user interface that enables end users to easily conduct and visualize the output of few-shot learning models. This interface is lightweight and can be hosted locally or on the web, providing the opportunity to reproducibly conduct, share, and crowd-source few-shot analyses.
A Topological-Framework to Improve Analysis of Machine Learning Model Performance
Jul 09, 2021

Abstract:As both machine learning models and the datasets on which they are evaluated have grown in size and complexity, the practice of using a few summary statistics to understand model performance has become increasingly problematic. This is particularly true in real-world scenarios where understanding model failure on certain subpopulations of the data is of critical importance. In this paper we propose a topological framework for evaluating machine learning models in which a dataset is treated as a "space" on which a model operates. This provides us with a principled way to organize information about model performance at both the global level (over the entire test set) and also the local level (on specific subpopulations). Finally, we describe a topological data structure, presheaves, which offer a convenient way to store and analyze model performance between different subpopulations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge