Ling Zhang
Missouri S&T EMC Laboratory, Rolla, MO, USA
SnapshotNet: Self-supervised Feature Learning for Point Cloud Data Segmentation Using Minimal Labeled Data
Jan 13, 2022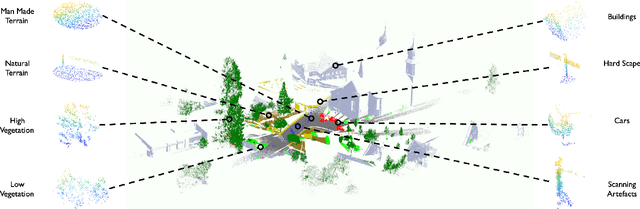
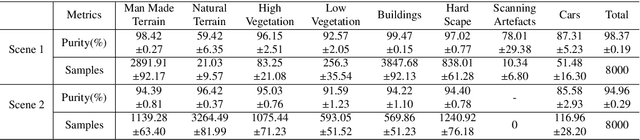
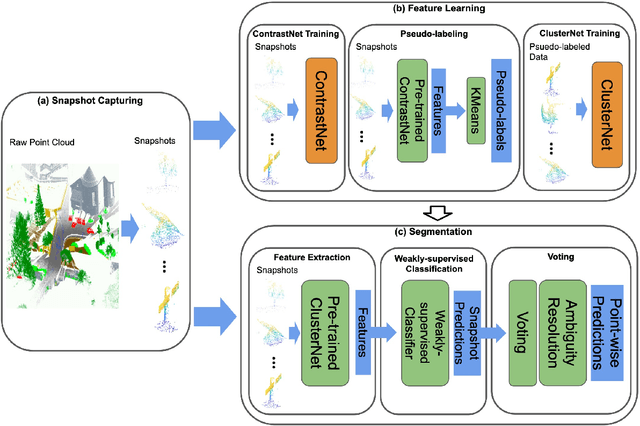
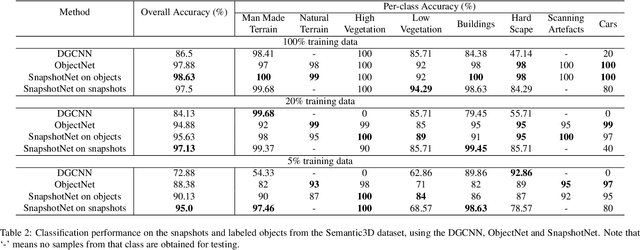
Abstract:Manually annotating complex scene point cloud datasets is both costly and error-prone. To reduce the reliance on labeled data, a new model called SnapshotNet is proposed as a self-supervised feature learning approach, which directly works on the unlabeled point cloud data of a complex 3D scene. The SnapshotNet pipeline includes three stages. In the snapshot capturing stage, snapshots, which are defined as local collections of points, are sampled from the point cloud scene. A snapshot could be a view of a local 3D scan directly captured from the real scene, or a virtual view of such from a large 3D point cloud dataset. Snapshots could also be sampled at different sampling rates or fields of view (FOVs), thus multi-FOV snapshots, to capture scale information from the scene. In the feature learning stage, a new pre-text task called multi-FOV contrasting is proposed to recognize whether two snapshots are from the same object or not, within the same FOV or across different FOVs. Snapshots go through two self-supervised learning steps: the contrastive learning step with both part and scale contrasting, followed by a snapshot clustering step to extract higher level semantic features. Then a weakly-supervised segmentation stage is implemented by first training a standard SVM classifier on the learned features with a small fraction of labeled snapshots. The trained SVM is used to predict labels for input snapshots and predicted labels are converted into point-wise label assignments for semantic segmentation of the entire scene using a voting procedure. The experiments are conducted on the Semantic3D dataset and the results have shown that the proposed method is capable of learning effective features from snapshots of complex scene data without any labels. Moreover, the proposed method has shown advantages when comparing to the SOA method on weakly-supervised point cloud semantic segmentation.
A free lunch from ViT:Adaptive Attention Multi-scale Fusion Transformer for Fine-grained Visual Recognition
Oct 11, 2021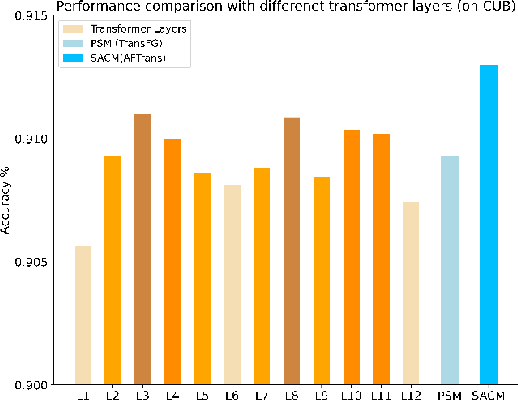
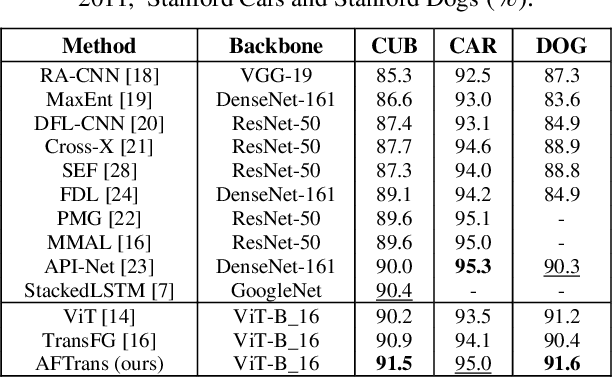
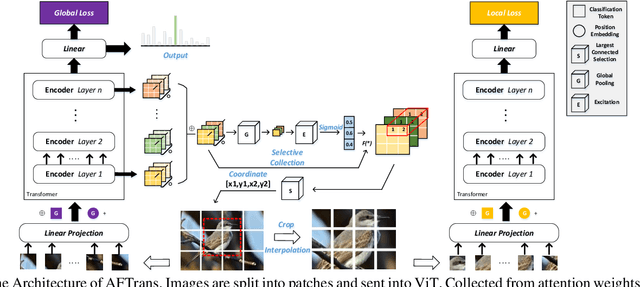
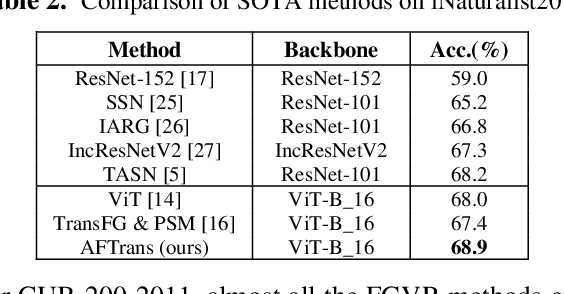
Abstract:Learning subtle representation about object parts plays a vital role in fine-grained visual recognition (FGVR) field. The vision transformer (ViT) achieves promising results on computer vision due to its attention mechanism. Nonetheless, with the fixed size of patches in ViT, the class token in deep layer focuses on the global receptive field and cannot generate multi-granularity features for FGVR. To capture region attention without box annotations and compensate for ViT shortcomings in FGVR, we propose a novel method named Adaptive attention multi-scale Fusion Transformer (AFTrans). The Selective Attention Collection Module (SACM) in our approach leverages attention weights in ViT and filters them adaptively to correspond with the relative importance of input patches. The multiple scales (global and local) pipeline is supervised by our weights sharing encoder and can be easily trained end-to-end. Comprehensive experiments demonstrate that AFTrans can achieve SOTA performance on three published fine-grained benchmarks: CUB-200-2011, Stanford Dogs and iNat2017.
Learning to Solve the AC Optimal Power Flow via a Lagrangian Approach
Oct 04, 2021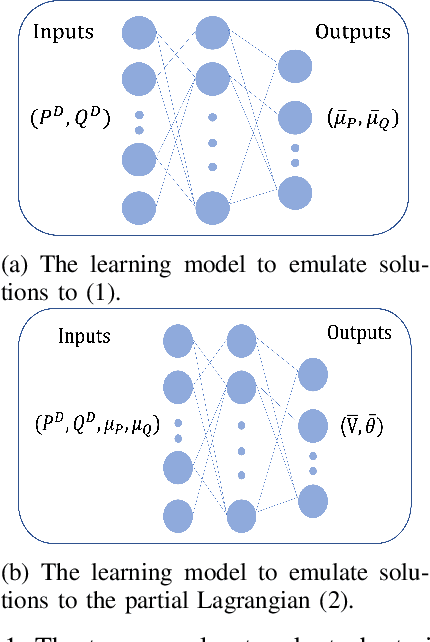
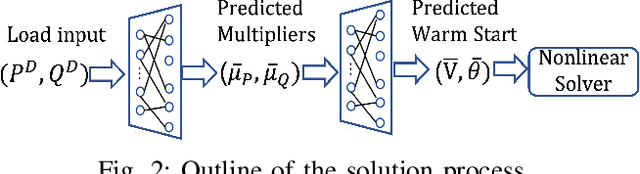
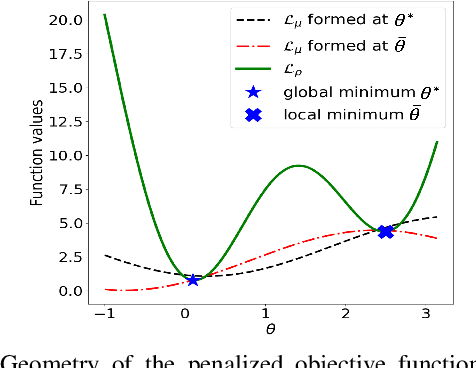
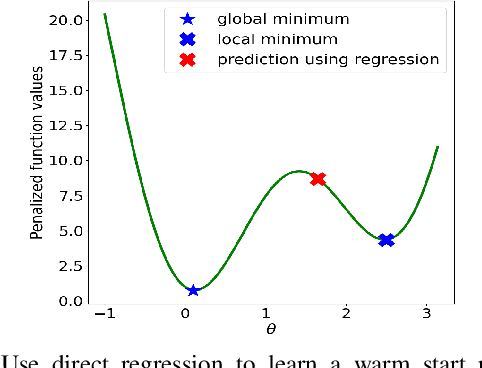
Abstract:Using deep neural networks to predict the solutions of AC optimal power flow (ACOPF) problems has been an active direction of research. However, because the ACOPF is nonconvex, it is difficult to construct a good data set that contains mostly globally optimal solutions. To overcome the challenge that the training data may contain suboptimal solutions, we propose a Lagrangian-based approach. First, we use a neural network to learn the dual variables of the ACOPF problem. Then we use a second neural network to predict solutions of the partial Lagrangian from the predicted dual variables. Since the partial Lagrangian has a much better optimization landscape, we use the predicted solutions from the neural network as a warm start for the ACOPF problem. Using standard and modified IEEE 22-bus, 39-bus, and 118-bus networks, we show that our approach is able to obtain the globally optimal cost even when the training data is mostly comprised of suboptimal solutions.
CANet: A Context-Aware Network for Shadow Removal
Aug 23, 2021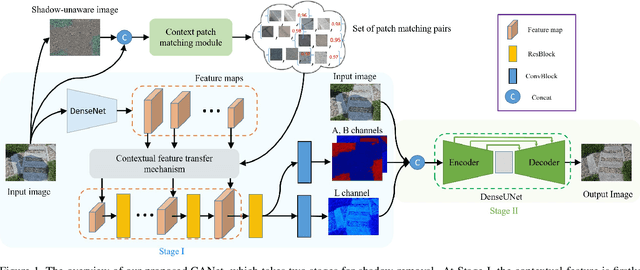
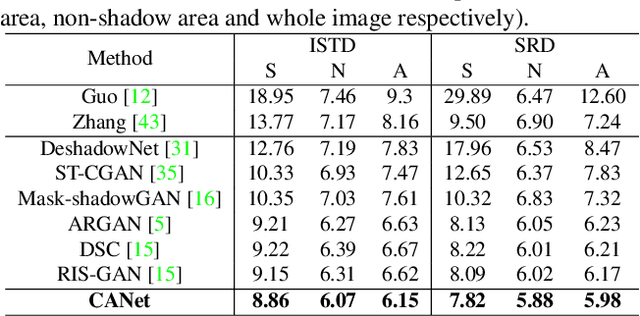
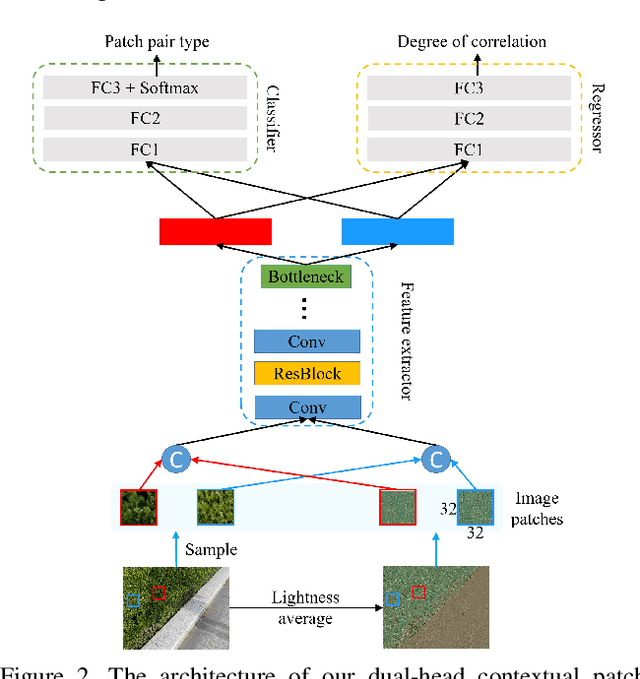
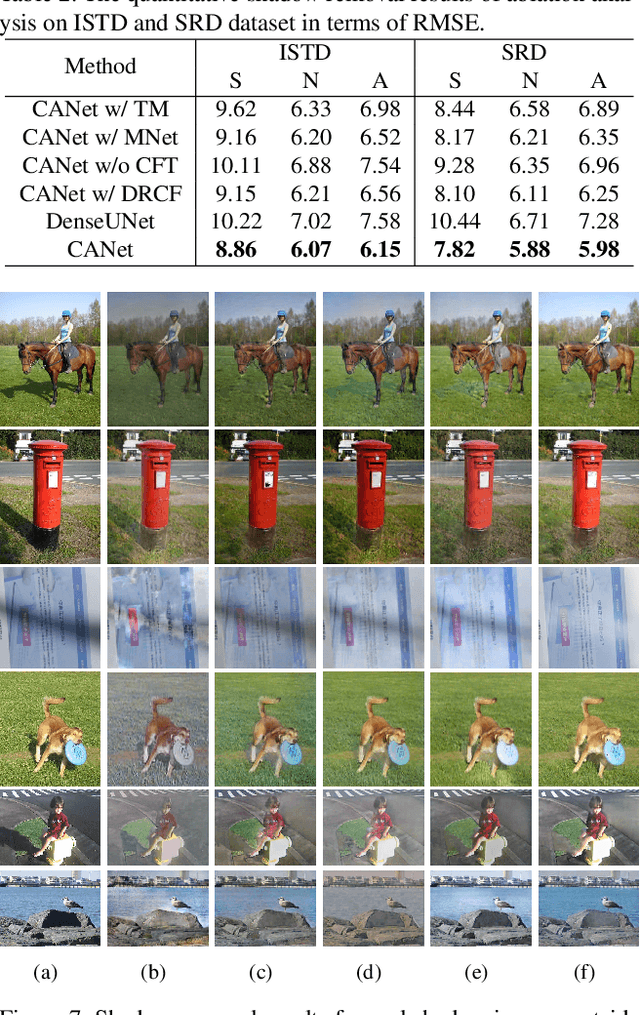
Abstract:In this paper, we propose a novel two-stage context-aware network named CANet for shadow removal, in which the contextual information from non-shadow regions is transferred to shadow regions at the embedded feature spaces. At Stage-I, we propose a contextual patch matching (CPM) module to generate a set of potential matching pairs of shadow and non-shadow patches. Combined with the potential contextual relationships between shadow and non-shadow regions, our well-designed contextual feature transfer (CFT) mechanism can transfer contextual information from non-shadow to shadow regions at different scales. With the reconstructed feature maps, we remove shadows at L and A/B channels separately. At Stage-II, we use an encoder-decoder to refine current results and generate the final shadow removal results. We evaluate our proposed CANet on two benchmark datasets and some real-world shadow images with complex scenes. Extensive experimental results strongly demonstrate the efficacy of our proposed CANet and exhibit superior performance to state-of-the-arts.
Distilling Neuron Spike with High Temperature in Reinforcement Learning Agents
Aug 05, 2021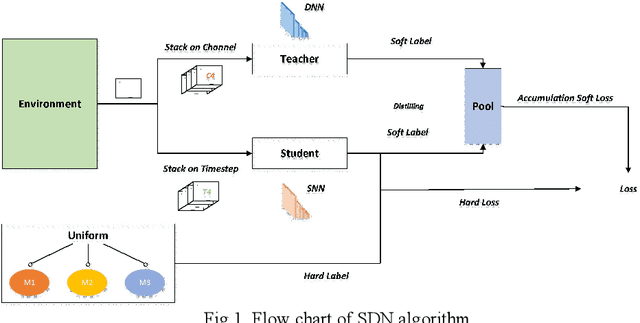
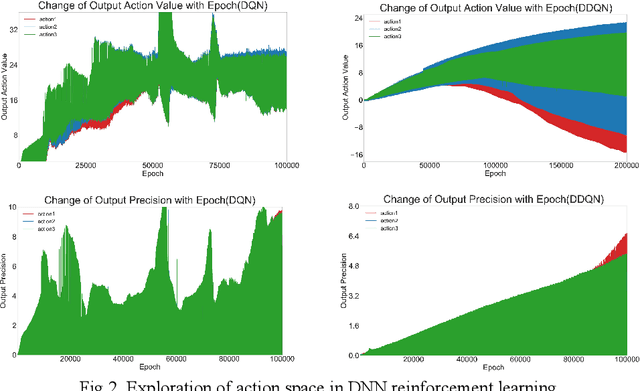
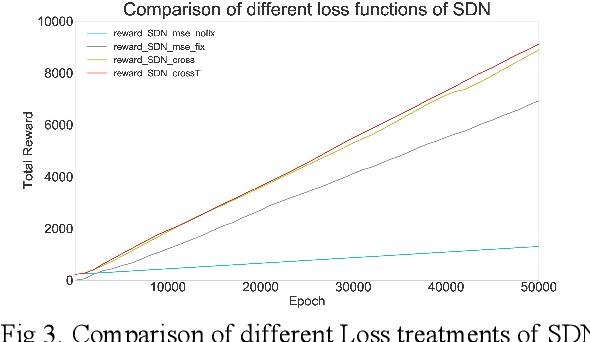
Abstract:Spiking neural network (SNN), compared with depth neural network (DNN), has faster processing speed, lower energy consumption and more biological interpretability, which is expected to approach Strong AI. Reinforcement learning is similar to learning in biology. It is of great significance to study the combination of SNN and RL. We propose the reinforcement learning method of spike distillation network (SDN) with STBP. This method uses distillation to effectively avoid the weakness of STBP, which can achieve SOTA performance in classification, and can obtain a smaller, faster convergence and lower power consumption SNN reinforcement learning model. Experiments show that our method can converge faster than traditional SNN reinforcement learning and DNN reinforcement learning methods, about 1000 epochs faster, and obtain SNN 200 times smaller than DNN. We also deploy SDN to the PKU nc64c chip, which proves that SDN has lower power consumption than DNN, and the power consumption of SDN is more than 600 times lower than DNN on large-scale devices. SDN provides a new way of SNN reinforcement learning, and can achieve SOTA performance, which proves the possibility of further development of SNN reinforcement learning.
Fast PDN Impedance Prediction Using Deep Learning
Jun 20, 2021
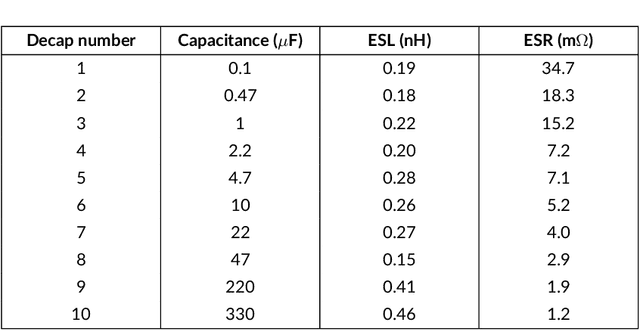
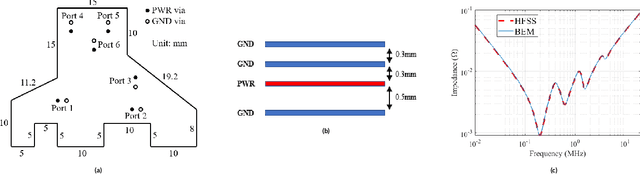

Abstract:Modeling and simulating a power distribution network (PDN) for printed circuit boards (PCBs) with irregular board shapes and multi-layer stackup is computationally inefficient using full-wave simulations. This paper presents a new concept of using deep learning for PDN impedance prediction. A boundary element method (BEM) is applied to efficiently calculate the impedance for arbitrary board shape and stackup. Then over one million boards with different shapes, stackup, IC location, and decap placement are randomly generated to train a deep neural network (DNN). The trained DNN can predict the impedance accurately for new board configurations that have not been used for training. The consumed time using the trained DNN is only 0.1 seconds, which is over 100 times faster than the BEM method and 5000 times faster than full-wave simulations.
Automated Discovery of Real-Time Network Camera Data From Heterogeneous Web Pages
Mar 23, 2021

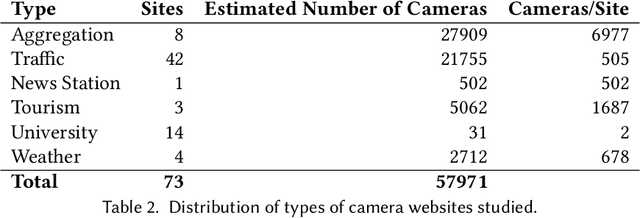
Abstract:Reduction in the cost of Network Cameras along with a rise in connectivity enables entities all around the world to deploy vast arrays of camera networks. Network cameras offer real-time visual data that can be used for studying traffic patterns, emergency response, security, and other applications. Although many sources of Network Camera data are available, collecting the data remains difficult due to variations in programming interface and website structures. Previous solutions rely on manually parsing the target website, taking many hours to complete. We create a general and automated solution for aggregating Network Camera data spread across thousands of uniquely structured web pages. We analyze heterogeneous web page structures and identify common characteristics among 73 sample Network Camera websites (each website has multiple web pages). These characteristics are then used to build an automated camera discovery module that crawls and aggregates Network Camera data. Our system successfully extracts 57,364 Network Cameras from 237,257 unique web pages.
3D Graph Anatomy Geometry-Integrated Network for Pancreatic Mass Segmentation, Diagnosis, and Quantitative Patient Management
Dec 08, 2020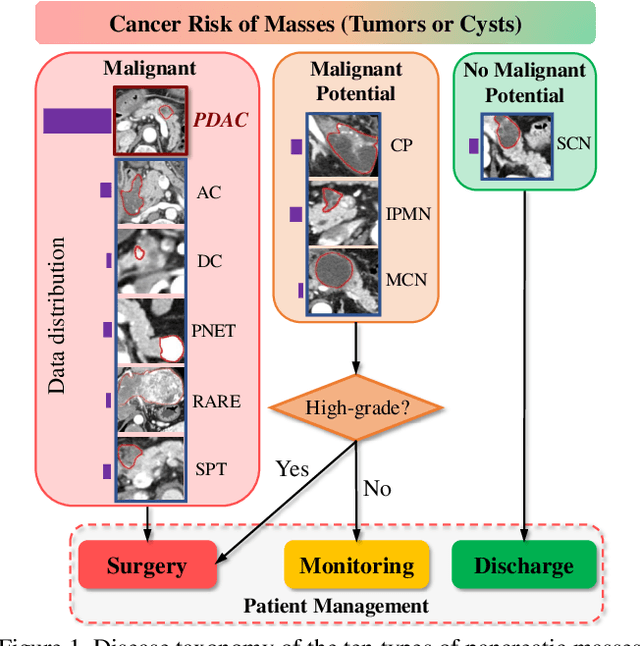
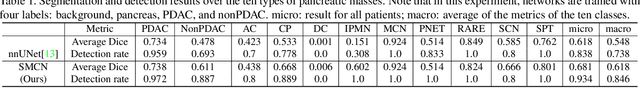
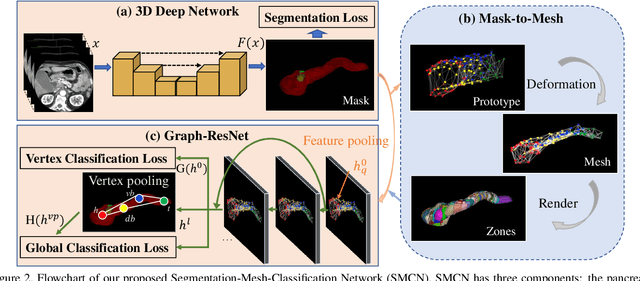
Abstract:The pancreatic disease taxonomy includes ten types of masses (tumors or cysts)[20,8]. Previous work focuses on developing segmentation or classification methods only for certain mass types. Differential diagnosis of all mass types is clinically highly desirable [20] but has not been investigated using an automated image understanding approach. We exploit the feasibility to distinguish pancreatic ductal adenocarcinoma (PDAC) from the nine other nonPDAC masses using multi-phase CT imaging. Both image appearance and the 3D organ-mass geometry relationship are critical. We propose a holistic segmentation-mesh-classification network (SMCN) to provide patient-level diagnosis, by fully utilizing the geometry and location information, which is accomplished by combining the anatomical structure and the semantic detection-by-segmentation network. SMCN learns the pancreas and mass segmentation task and builds an anatomical correspondence-aware organ mesh model by progressively deforming a pancreas prototype on the raw segmentation mask (i.e., mask-to-mesh). A new graph-based residual convolutional network (Graph-ResNet), whose nodes fuse the information of the mesh model and feature vectors extracted from the segmentation network, is developed to produce the patient-level differential classification results. Extensive experiments on 661 patients' CT scans (five phases per patient) show that SMCN can improve the mass segmentation and detection accuracy compared to the strong baseline method nnUNet (e.g., for nonPDAC, Dice: 0.611 vs. 0.478; detection rate: 89% vs. 70%), achieve similar sensitivity and specificity in differentiating PDAC and nonPDAC as expert radiologists (i.e., 94% and 90%), and obtain results comparable to a multimodality test [20] that combines clinical, imaging, and molecular testing for clinical management of patients.
DeepPrognosis: Preoperative Prediction of Pancreatic Cancer Survival and Surgical Margin via Contrast-Enhanced CT Imaging
Aug 26, 2020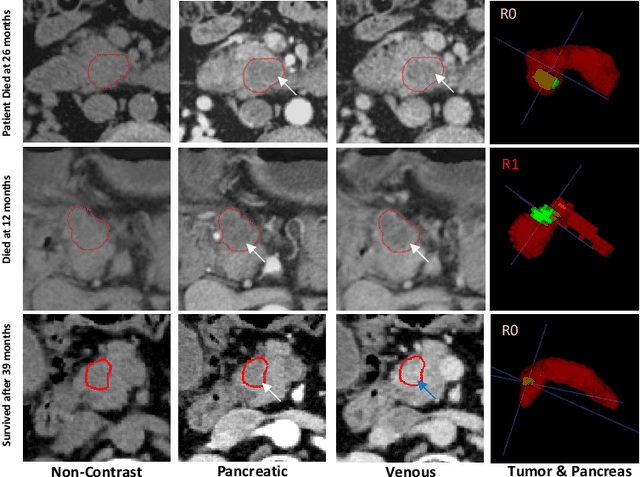
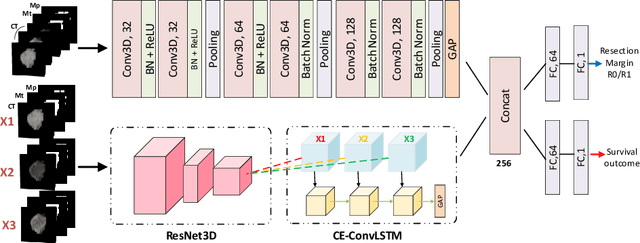
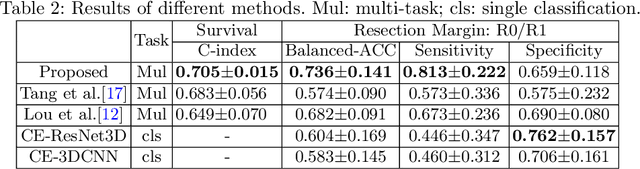
Abstract:Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers and carries a dismal prognosis. Surgery remains the best chance of a potential cure for patients who are eligible for initial resection of PDAC. However, outcomes vary significantly even among the resected patients of the same stage and received similar treatments. Accurate preoperative prognosis of resectable PDACs for personalized treatment is thus highly desired. Nevertheless, there are no automated methods yet to fully exploit the contrast-enhanced computed tomography (CE-CT) imaging for PDAC. Tumor attenuation changes across different CT phases can reflect the tumor internal stromal fractions and vascularization of individual tumors that may impact the clinical outcomes. In this work, we propose a novel deep neural network for the survival prediction of resectable PDAC patients, named as 3D Contrast-Enhanced Convolutional Long Short-Term Memory network(CE-ConvLSTM), which can derive the tumor attenuation signatures or patterns from CE-CT imaging studies. We present a multi-task CNN to accomplish both tasks of outcome and margin prediction where the network benefits from learning the tumor resection margin related features to improve survival prediction. The proposed framework can improve the prediction performances compared with existing state-of-the-art survival analysis approaches. The tumor signature built from our model has evidently added values to be combined with the existing clinical staging system.
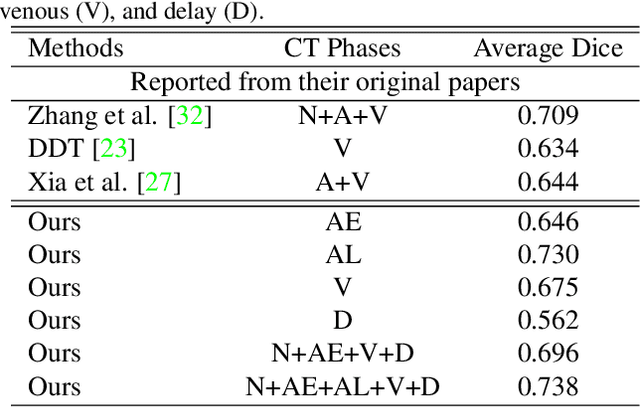
Robust Pancreatic Ductal Adenocarcinoma Segmentation with Multi-Institutional Multi-Phase Partially-Annotated CT Scans
Aug 24, 2020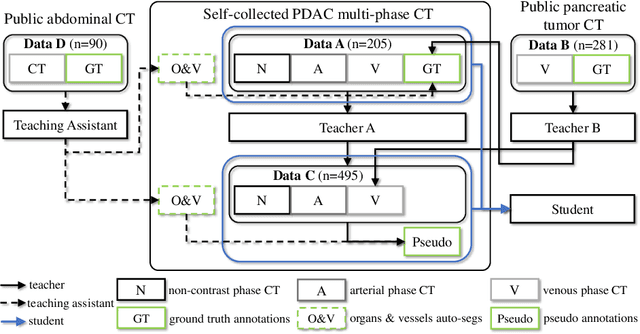
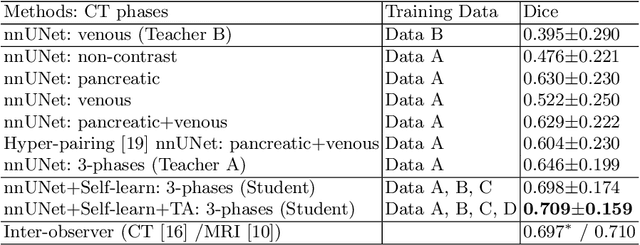
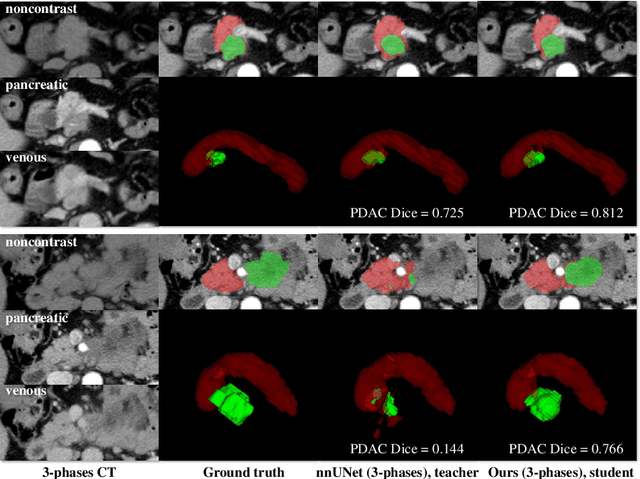
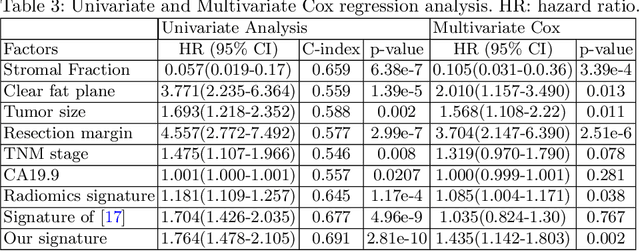
Abstract:Accurate and automated tumor segmentation is highly desired since it has the great potential to increase the efficiency and reproducibility of computing more complete tumor measurements and imaging biomarkers, comparing to (often partial) human measurements. This is probably the only viable means to enable the large-scale clinical oncology patient studies that utilize medical imaging. Deep learning approaches have shown robust segmentation performances for certain types of tumors, e.g., brain tumors in MRI imaging, when a training dataset with plenty of pixel-level fully-annotated tumor images is available. However, more than often, we are facing the challenge that only (very) limited annotations are feasible to acquire, especially for hard tumors. Pancreatic ductal adenocarcinoma (PDAC) segmentation is one of the most challenging tumor segmentation tasks, yet critically important for clinical needs. Previous work on PDAC segmentation is limited to the moderate amounts of annotated patient images (n<300) from venous or venous+arterial phase CT scans. Based on a new self-learning framework, we propose to train the PDAC segmentation model using a much larger quantity of patients (n~=1,000), with a mix of annotated and un-annotated venous or multi-phase CT images. Pseudo annotations are generated by combining two teacher models with different PDAC segmentation specialties on unannotated images, and can be further refined by a teaching assistant model that identifies associated vessels around the pancreas. A student model is trained on both manual and pseudo annotated multi-phase images. Experiment results show that our proposed method provides an absolute improvement of 6.3% Dice score over the strong baseline of nnUNet trained on annotated images, achieving the performance (Dice = 0.71) similar to the inter-observer variability between radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge