Leo Joskowicz
A multi-centre, multi-device benchmark dataset for landmark-based comprehensive fetal biometry
Dec 19, 2025Abstract:Accurate fetal growth assessment from ultrasound (US) relies on precise biometry measured by manually identifying anatomical landmarks in standard planes. Manual landmarking is time-consuming, operator-dependent, and sensitive to variability across scanners and sites, limiting the reproducibility of automated approaches. There is a need for multi-source annotated datasets to develop artificial intelligence-assisted fetal growth assessment methods. To address this bottleneck, we present an open, multi-centre, multi-device benchmark dataset of fetal US images with expert anatomical landmark annotations for clinically used fetal biometric measurements. These measurements include head bi-parietal and occipito-frontal diameters, abdominal transverse and antero-posterior diameters, and femoral length. The dataset comprises 4,513 de-identified US images from 1,904 subjects acquired at three clinical sites using seven different US devices. We provide standardised, subject-disjoint train/test splits, evaluation code, and baseline results to enable fair and reproducible comparison of methods. Using an automatic biometry model, we quantify domain shift and demonstrate that training and evaluation confined to a single centre substantially overestimate performance relative to multi-centre testing. To the best of our knowledge, this is the first publicly available multi-centre, multi-device, landmark-annotated dataset that covers all primary fetal biometry measures, providing a robust benchmark for domain adaptation and multi-centre generalisation in fetal biometry and enabling more reliable AI-assisted fetal growth assessment across centres. All data, annotations, training code, and evaluation pipelines are made publicly available.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
SegQC: a segmentation network-based framework for multi-metric segmentation quality control and segmentation error detection in volumetric medical images
Nov 12, 2024Abstract:Quality control of structures segmentation in volumetric medical images is important for identifying segmentation errors in clinical practice and for facilitating model development. This paper introduces SegQC, a novel framework for segmentation quality estimation and segmentation error detection. SegQC computes an estimate measure of the quality of a segmentation in volumetric scans and in their individual slices and identifies possible segmentation error regions within a slice. The key components include: 1. SegQC-Net, a deep network that inputs a scan and its segmentation mask and outputs segmentation error probabilities for each voxel in the scan; 2. three new segmentation quality metrics, two overlap metrics and a structure size metric, computed from the segmentation error probabilities; 3. a new method for detecting possible segmentation errors in scan slices computed from the segmentation error probabilities. We introduce a new evaluation scheme to measure segmentation error discrepancies based on an expert radiologist corrections of automatically produced segmentations that yields smaller observer variability and is closer to actual segmentation errors. We demonstrate SegQC on three fetal structures in 198 fetal MRI scans: fetal brain, fetal body and the placenta. To assess the benefits of SegQC, we compare it to the unsupervised Test Time Augmentation (TTA)-based quality estimation. Our studies indicate that SegQC outperforms TTA-based quality estimation in terms of Pearson correlation and MAE for fetal body and fetal brain structures segmentation. Our segmentation error detection method achieved recall and precision rates of 0.77 and 0.48 for fetal body, and 0.74 and 0.55 for fetal brain segmentation error detection respectively. SegQC enhances segmentation metrics estimation for whole scans and individual slices, as well as provides error regions detection.
Test-time augmentation-based active learning and self-training for label-efficient segmentation
Aug 21, 2023



Abstract:Deep learning techniques depend on large datasets whose annotation is time-consuming. To reduce annotation burden, the self-training (ST) and active-learning (AL) methods have been developed as well as methods that combine them in an iterative fashion. However, it remains unclear when each method is the most useful, and when it is advantageous to combine them. In this paper, we propose a new method that combines ST with AL using Test-Time Augmentations (TTA). First, TTA is performed on an initial teacher network. Then, cases for annotation are selected based on the lowest estimated Dice score. Cases with high estimated scores are used as soft pseudo-labels for ST. The selected annotated cases are trained with existing annotated cases and ST cases with border slices annotations. We demonstrate the method on MRI fetal body and placenta segmentation tasks with different data variability characteristics. Our results indicate that ST is highly effective for both tasks, boosting performance for in-distribution (ID) and out-of-distribution (OOD) data. However, while self-training improved the performance of single-sequence fetal body segmentation when combined with AL, it slightly deteriorated performance of multi-sequence placenta segmentation on ID data. AL was helpful for the high variability placenta data, but did not improve upon random selection for the single-sequence body data. For fetal body segmentation sequence transfer, combining AL with ST following ST iteration yielded a Dice of 0.961 with only 6 original scans and 2 new sequence scans. Results using only 15 high-variability placenta cases were similar to those using 50 cases. Code is available at: https://github.com/Bella31/TTA-quality-estimation-ST-AL
Simultaneous column-based deep learning progression analysis of atrophy associated with AMD in longitudinal OCT studies
Jul 31, 2023



Abstract:Purpose: Disease progression of retinal atrophy associated with AMD requires the accurate quantification of the retinal atrophy changes on longitudinal OCT studies. It is based on finding, comparing, and delineating subtle atrophy changes on consecutive pairs (prior and current) of unregistered OCT scans. Methods: We present a fully automatic end-to-end pipeline for the simultaneous detection and quantification of time-related atrophy changes associated with dry AMD in pairs of OCT scans of a patient. It uses a novel simultaneous multi-channel column-based deep learning model trained on registered pairs of OCT scans that concurrently detects and segments retinal atrophy segments in consecutive OCT scans by classifying light scattering patterns in matched pairs of vertical pixel-wide columns (A-scans) in registered prior and current OCT slices (B-scans). Results: Experimental results on 4,040 OCT slices with 5.2M columns from 40 scans pairs of 18 patients (66% training/validation, 33% testing) with 24.13+-14.0 months apart in which Complete RPE and Outer Retinal Atrophy (cRORA) was identified in 1,998 OCT slices (735 atrophy lesions from 3,732 segments, 0.45M columns) yield a mean atrophy segments detection precision, recall of 0.90+-0.09, 0.95+-0.06 and 0.74+-0.18, 0.94+-0.12 for atrophy lesions with AUC=0.897, all above observer variability. Simultaneous classification outperforms standalone classification precision and recall by 30+-62% and 27+-0% for atrophy segments and lesions. Conclusions: simultaneous column-based detection and quantification of retinal atrophy changes associated with AMD is accurate and outperforms standalone classification methods. Translational relevance: an automatic and efficient way to detect and quantify retinal atrophy changes associated with AMD.
Contour Dice loss for structures with Fuzzy and Complex Boundaries in Fetal MRI
Sep 25, 2022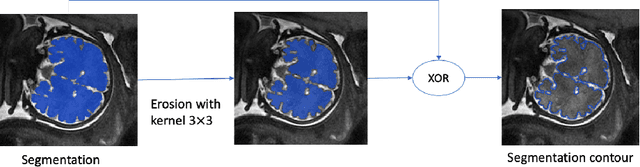
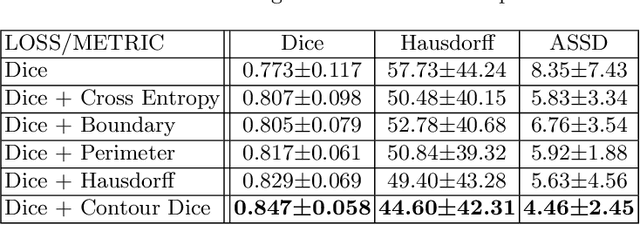
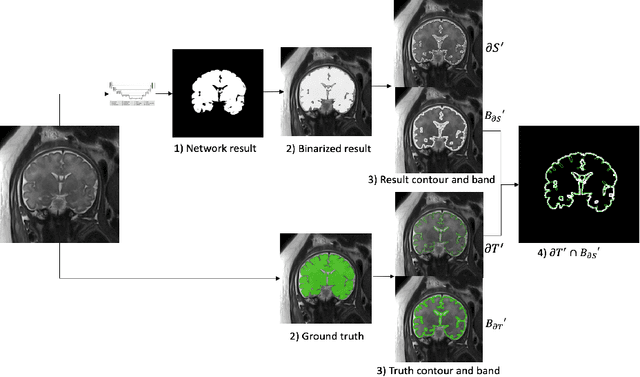
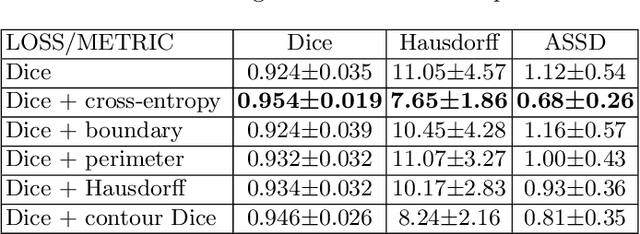
Abstract:Volumetric measurements of fetal structures in MRI are time consuming and error prone and therefore require automatic segmentation. Placenta segmentation and accurate fetal brain segmentation for gyrification assessment are particularly challenging because of the placenta fuzzy boundaries and the fetal brain cortex complex foldings. In this paper, we study the use of the Contour Dice loss for both problems and compare it to other boundary losses and to the combined Dice and Cross-Entropy loss. The loss is computed efficiently for each slice via erosion, dilation and XOR operators. We describe a new formulation of the loss akin to the Contour Dice metric. The combination of the Dice loss and the Contour Dice yielded the best performance for placenta segmentation. For fetal brain segmentation, the best performing loss was the combined Dice with Cross-Entropy loss followed by the Dice with Contour Dice loss, which performed better than other boundary losses.
Partial annotations for the segmentation of large structures with low annotation cost
Sep 25, 2022Abstract:Deep learning methods have been shown to be effective for the automatic segmentation of structures and pathologies in medical imaging. However, they require large annotated datasets, whose manual segmentation is a tedious and time-consuming task, especially for large structures. We present a new method of partial annotations that uses a small set of consecutive annotated slices from each scan with an annotation effort that is equal to that of only few annotated cases. The training with partial annotations is performed by using only annotated blocks, incorporating information about slices outside the structure of interest and modifying a batch loss function to consider only the annotated slices. To facilitate training in a low data regime, we use a two-step optimization process. We tested the method with the popular soft Dice loss for the fetal body segmentation task in two MRI sequences, TRUFI and FIESTA, and compared full annotation regime to partial annotations with a similar annotation effort. For TRUFI data, the use of partial annotations yielded slightly better performance on average compared to full annotations with an increase in Dice score from 0.936 to 0.942, and a substantial decrease in Standard Deviations (STD) of Dice score by 22% and Average Symmetric Surface Distance (ASSD) by 15%. For the FIESTA sequence, partial annotations also yielded a decrease in STD of the Dice score and ASSD metrics by 27.5% and 33% respectively for in-distribution data, and a substantial improvement also in average performance on out-of-distribution data, increasing Dice score from 0.84 to 0.9 and decreasing ASSD from 7.46 to 4.01 mm. The two-step optimization process was helpful for partial annotations for both in-distribution and out-of-distribution data. The partial annotations method with the two-step optimizer is therefore recommended to improve segmentation performance under low data regime.
* 10 pages, 4 figures
Automatic fetal fat quantification from MRI
Sep 08, 2022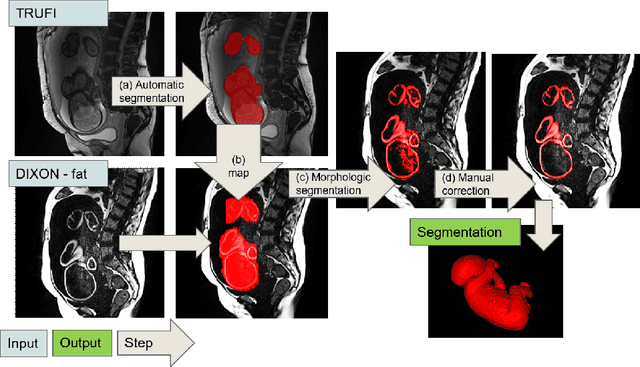
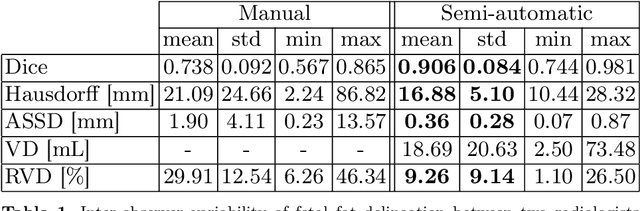
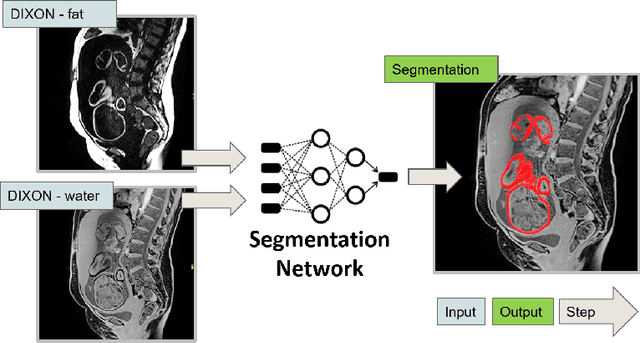
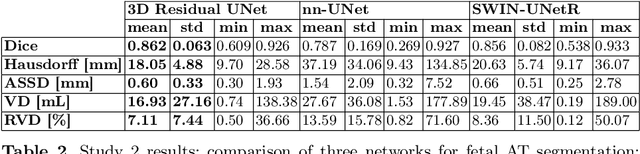
Abstract:Normal fetal adipose tissue (AT) development is essential for perinatal well-being. AT, or simply fat, stores energy in the form of lipids. Malnourishment may result in excessive or depleted adiposity. Although previous studies showed a correlation between the amount of AT and perinatal outcome, prenatal assessment of AT is limited by lacking quantitative methods. Using magnetic resonance imaging (MRI), 3D fat- and water-only images of the entire fetus can be obtained from two point Dixon images to enable AT lipid quantification. This paper is the first to present a methodology for developing a deep learning based method for fetal fat segmentation based on Dixon MRI. It optimizes radiologists' manual fetal fat delineation time to produce annotated training dataset. It consists of two steps: 1) model-based semi-automatic fetal fat segmentations, reviewed and corrected by a radiologist; 2) automatic fetal fat segmentation using DL networks trained on the resulting annotated dataset. Three DL networks were trained. We show a significant improvement in segmentation times (3:38 hours to < 1 hour) and observer variability (Dice of 0.738 to 0.906) compared to manual segmentation. Automatic segmentation of 24 test cases with the 3D Residual U-Net, nn-UNet and SWIN-UNetR transformer networks yields a mean Dice score of 0.863, 0.787 and 0.856, respectively. These results are better than the manual observer variability, and comparable to automatic adult and pediatric fat segmentation. A radiologist reviewed and corrected six new independent cases segmented using the best performing network, resulting in a Dice score of 0.961 and a significantly reduced correction time of 15:20 minutes. Using these novel segmentation methods and short MRI acquisition time, whole body subcutaneous lipids can be quantified for individual fetuses in the clinic and large-cohort research.
BiometryNet: Landmark-based Fetal Biometry Estimation from Standard Ultrasound Planes
Jun 29, 2022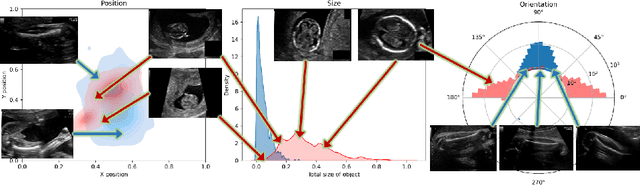
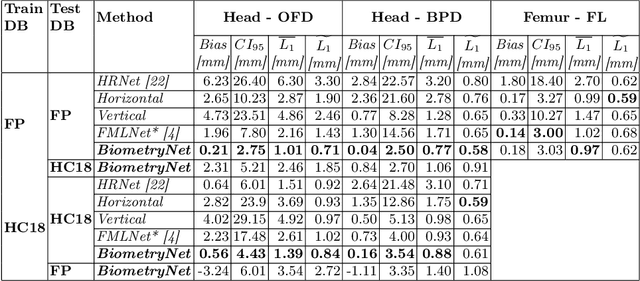
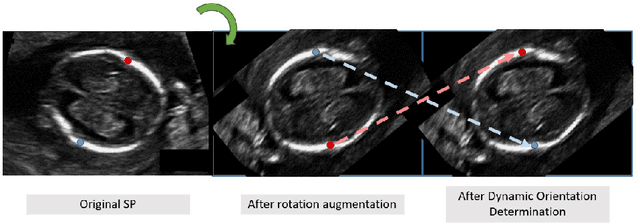
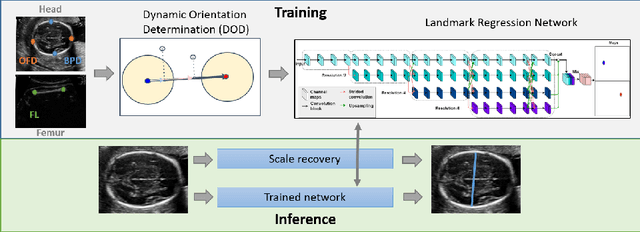
Abstract:Fetal growth assessment from ultrasound is based on a few biometric measurements that are performed manually and assessed relative to the expected gestational age. Reliable biometry estimation depends on the precise detection of landmarks in standard ultrasound planes. Manual annotation can be time-consuming and operator dependent task, and may results in high measurements variability. Existing methods for automatic fetal biometry rely on initial automatic fetal structure segmentation followed by geometric landmark detection. However, segmentation annotations are time-consuming and may be inaccurate, and landmark detection requires developing measurement-specific geometric methods. This paper describes BiometryNet, an end-to-end landmark regression framework for fetal biometry estimation that overcomes these limitations. It includes a novel Dynamic Orientation Determination (DOD) method for enforcing measurement-specific orientation consistency during network training. DOD reduces variabilities in network training, increases landmark localization accuracy, thus yields accurate and robust biometric measurements. To validate our method, we assembled a dataset of 3,398 ultrasound images from 1,829 subjects acquired in three clinical sites with seven different ultrasound devices. Comparison and cross-validation of three different biometric measurements on two independent datasets shows that BiometryNet is robust and yields accurate measurements whose errors are lower than the clinically permissible errors, outperforming other existing automated biometry estimation methods. Code is available at https://github.com/netanellavisdris/fetalbiometry.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022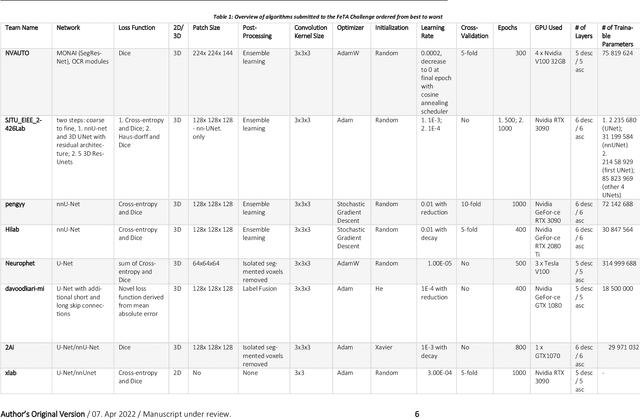
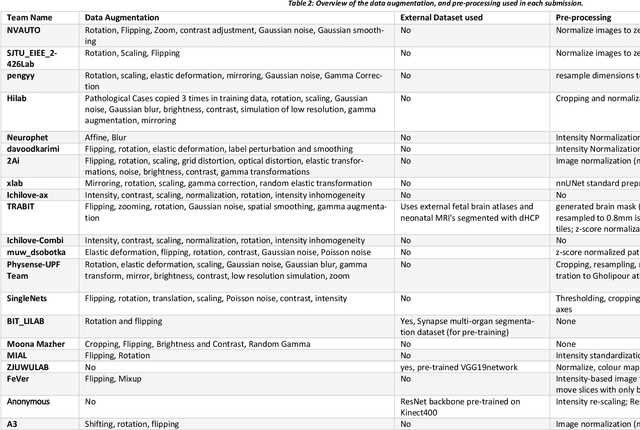
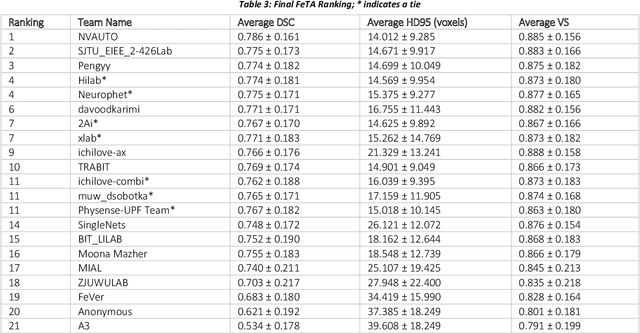
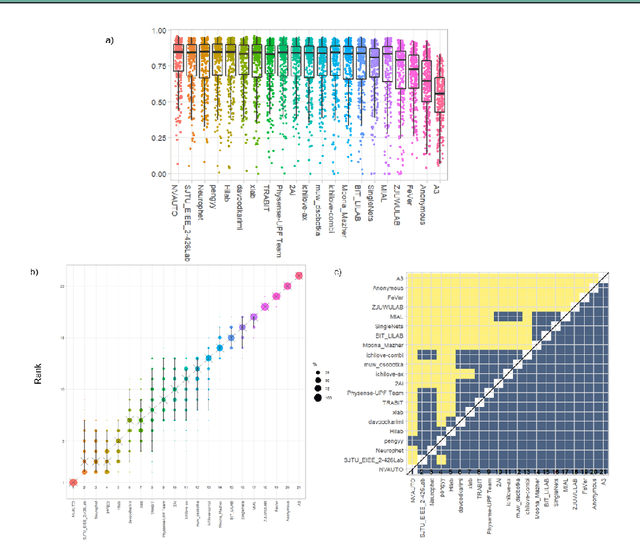
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge