Yuchen Pei
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025



Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022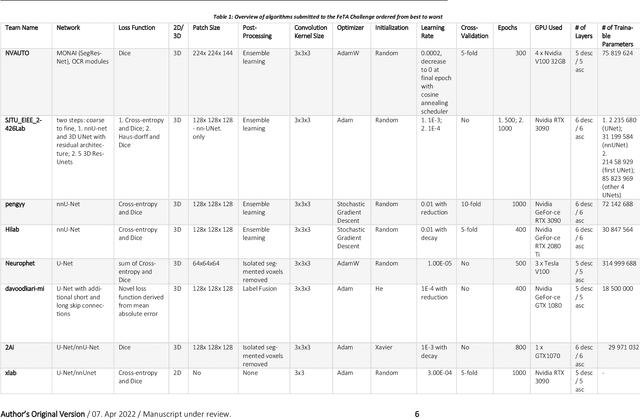
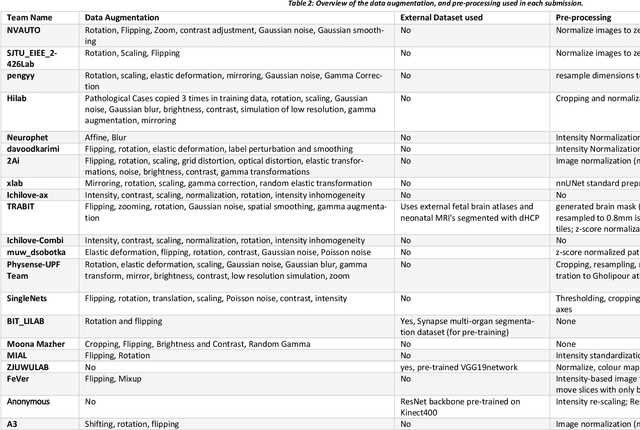
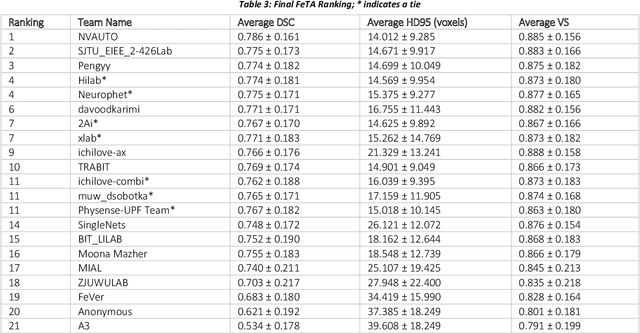
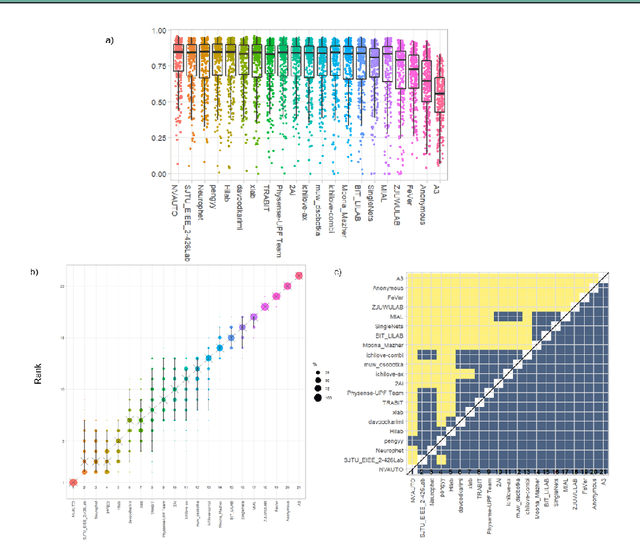
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge